Published online Mar 21, 2011. doi: 10.3748/wjg.v17.i11.1434
Revised: December 16, 2010
Accepted: December 23, 2010
Published online: March 21, 2011
AIM: To detect the proteomic variabilities of pancreatic intraepithelial neoplasia (PanIN) and pancreatic carcinoma (PC) induced by 7,12-dimethylbenzanthracene (DMBA) in rat models and to identify potential biomarkers.
METHODS: Sixty adult male Sprague Dawley rats were randomized into three groups. The rats had DMBA implanted into their pancreas for one (n = 20) or two months (n = 20) or assigned to the normal group (n = 20). The rats were killed after one or two months, and were evaluated histopathologically. Three tissue samples from each group of rats with either normal pancreas, PanIN (PanIN-2) or PC were examined by 2D-DIGE. The different expression spot features were analyzed by matrix-assisted laser desorption/ionization-time of flight/time of flight (MALDI-TOF/TOF) tandem mass spectrometry. The expression of enolase 1, a differentially expressed protein, was identified by immunohistochemistry.
RESULTS: There was significant difference in the proportions of neoplastic changes between the 1- and 2-mogroups (P = 0.0488). There was an increase in the frequency of adenocarcinomas in the 2-mo group compared with the 1-mo group (P = 0.0309). No neoplastic changes were observed in any of the animals in the normal group. Enolase 1, pancreatic ELA3B, necdin, Hbp23, CHD3, hnRNP A2/B1, Rap80, and Gnb2l1 were up-regulated in the PanIN and PC tissues, and CEL, TPT1, NME2, PCK2, an unnamed protein product, and glycine C-acetyltransferase were down-regulated in the PanIN and PC tissues. The immunohistochemical results showed that enolase 1 expression was up-regulated in the pancreatic cancer tissues of rats and humans.
CONCLUSION: The pancreatic protein expression changes induced by DMBA suggest potential molecular targets for the early diagnosis and treatment of PC.
- Citation: Wang L, Liu HL, Li Y, Yuan P. Proteomic analysis of pancreatic intraepithelial neoplasia and pancreatic carcinoma in rat models. World J Gastroenterol 2011; 17(11): 1434-1441
- URL: https://www.wjgnet.com/1007-9327/full/v17/i11/1434.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i11.1434
Pancreatic carcinoma (PC) is one of the most lethal human cancers, and 80%-90% of PCs are pancreatic ductal adenocarcinomas. Pancreatic cancer is characterized by a late presentation, and for all stages of this cancer, the five-year survival rate is less than 5%. It is currently the fourth most common cause of cancer death for both men and women[1]. This poor prognosis relates to the uniformly advanced disease stage at the time of diagnosis and its profound resistance to the existing therapies. An early detection of pancreatic cancer is required to improve its poor prognosis.
As the clinical diagnosis of pancreatic cancer is mostly at advanced stages, to study the mechanisms of PC and screen tumors for early diagnostic markers, it is important to establish animal models of precancerous pancreatic lesions and the early stages of PC with pathological characteristics resembling that of human PC. Pancreatic intraepithelial neoplasia (PanIN) has been identified as the precursor of pancreatic ductal adenocarcinoma[2,3], and animal models of PanIN and pancreatic ductal adenocarcinoma can provide opportunities to investigate earlier pancreatic cancers that are rare in human samples. Animal models involve successful chemical carcinogens such as 7,12-dimethylbenzanthracene (DMBA). DMBA implantation into the pancreas is known to induce ductal PanIN and PC in mice and rats through pathways that consistently involve K-ras gene mutations and the activation of Notch signaling, as in human pancreatic cancer[4-10]. At present, DMBA-induced pancreatic carcinogenesis models have been used in the identification of possible promoters or suppressors of PC[8,11,12], stable isotope glucose-tracer studies[13] and molecular analyses[10].
Proteomic techniques are emerging as important tools for studying the mechanisms of disease and finding potential biomarkers and new therapeutic targets, which enable investigators to determine whether a particular protein level is increased or decreased when comparing two different conditions (e.g. a diseased state and a non-diseased state)[14,15]. Although proteomic studies of PC in human samples have been reported, most of the samples were acquired from advanced PC but not precancerous pancreatic lesions or the early stages of PC. The protein expression changes from normal pancreas to PanIN and early stages of PC remain unclear. In the present research, we studied the proteomic variabilities of rat models of DMBA-induced PanIN and PC using proteomic techniques, two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) and mass spectrometry, to reveal new potential biomarkers for PanIN and PC.
Sixty adult male Sprague Dawley (SD) rats (100-110 g) were randomized into three groups. The rats in the 1- (n = 20) and 2-mo groups (n = 20) had DMBA (10 mg/100 g) directly implanted into their pancreas, according to a previously established protocol[9], and the surviving rats were killed after one month and two months, respectively. A normal group of 20 rats was killed after two months. The carcinogen implant site was separated from the rest of the pancreas, the pancreatic nodules were fixed in formalin and embedded in paraffin, and multiple 5-mm sections were prepared and stained with hematoxylin and eosin for routine histological examinations. The pancreas slides were reviewed by a single pathologist and evaluated histologically according to the PanIN classification system[2,3].
Three rat models with either normal pancreas (group A), PanIN (group B, PanIN-2), or PC (group C) were established, and three samples per group were collected. Approximately 300 mg of each tissue sample was divided into 3-mm3 pieces and then homogenized with a Dounce’s homogenizer on ice in 1 mL lysis solution containing 7 mol/L urea, 2 mol/L thiourea, 4% CHAPS and a protease inhibitor mixture (Bio-Rad). The sample lysates were then placed into Eppendorf tubes and sonicated on ice for 10 s, and this procedure was repeated eight times. The sample lysates were centrifuged at 14 000 rpm for 60 min, and the supernatants were collected. The total protein concentration in each sample was determined by the Bio-Rad method.
The protein concentration in each sample was dissolved to 5 μg/μL with lysis buffer, and equal amounts of protein from each individual sample were pooled to make the internal standard. The pH of all the samples and the internal standard was adjusted to 8.0-9.0. Five DIGE gels were included in the experimental design, as shown in Table 1. Two different samples in each gel contained 50 μg protein labeled with 1 μL green (Cy3) or red (Cy5) fluorescent dyes (Amersham Biosciences). The third sample contained 50 μg internal standard labeled with yellow (Cy2) fluorescent dye. The labeling reaction was performed on ice for 30 min in the dark and quenched with 1 μL of 10 mmol/L lysine for 10 min.
| Gel | Cy2 | Cy3 | Cy5 |
| Gel1 | Internal standard | A1 | B3 |
| Gel2 | Internal standard | B3 | C2 |
| Gel3 | Internal standard | C1 | A2 |
| Gel4 | Internal standard | B2 | C3 |
| Gel5 | Internal standard | A3 | B1 |
One-dimensional isoelectric focusing was carried out on an Amersham Biosciences Ettan IPGphor IEF system. The samples (100 μg) were loaded using 13-cm pH-3-10 IPG strips. Isoelectric focusing was performed at 30 V for 12 h, 500 V for 1 h, 1000 V for 1 h, 8000 V for 8 h and 500 V for 4 h. After IEF, the IPG strips were equilibrated with an equilibration buffer containing 50 mmol/L Tris-HCl, 6 mol/L urea, 30% glycerol, 2% SDS, and 1% DTT for 15 min at room temperature, followed by 2.5% iodoacetamide instead of 1% DTT in equilibration buffer for another 15 min. The equilibrated strip was transferred to the top of 1 mm of a 12.5% SDS-polyacrylamide gel and fixed with 0.5% agarose. The second dimension of SDS-PAGE was performed at a constant current of 15 mA/gel for 30 min and 30 mA/gel until the bromphenol blue fronts reached 0.5 cm from the bottom of the gel.
The fluorescent dye-labeled proteins in each gel were scanned with a Typhoon 9400 fluorescence scanner (Amersham Biosciences) at different wavelengths specific for Cy2 (488/520 nm), Cy3 (532/580 nm) and Cy5 (633/670 nm). Image analysis of 2D-DIGE was performed with Decyder 5.0 software (GE Healthcare) according to the manufacturer’s recommendations. Differential protein analysis of the three groups was carried out using one-way ANOVA. Differentially expressed protein spots (P < 0.01) were marked.
Two-dimensional electrophoresis of preparative gels containing 1 mg protein was performed like 2D-DIGE, and the gels were stained with Coomassie brilliant blue. The protein spots to be identified were manually excised from the preparative gels. The gel pieces were de-stained in 3% acetic acid and 100 mmol/L ammonium bicarbonate, and in-gel digestion was performed for 20 h at 37°C in 40 mmol/L ammonium bicarbonate containing 0.01 μg/μL trypsin. The tryptic peptides were extracted with 0.1% TFA (60% ACN from the supernatants), and the extracts were lyophilized and saved at -80°C.
The lyophilized peptide mixtures were re-dissolved in 0.1% TFA, the peptide solution was washed with 0.1% TFA, and the 60% ACN was mixed with HCCA (5 mg/mL). Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) and MALDI-TOF/TOF mass spectrometry were obtained using a Bruker-Daltonics Autoflex TOF-TOF Lift mass spectrometer. Peptide mass fingerprints (PMFs) were acquired for each differentially expressed protein in positive-reflection mode (20 kV accelerating voltage, 23 kV reflecting voltage). The PMFs were searched in the database of the National Center for Biotechnology Information (NCBI) of Rattus using the software Mascot. Carbamidomethyl was specified as fixed modification, and the oxidation as variable modifications, and the peptide mass tolerance was ± 100 ppm. The statistically significant protein scores were found (P < 0.05), and, if more than one protein was identified, the single protein with the top score was matched to the protein spot.
Three pairs of rat normal pancreas and DMBA-induced PC tissues and 30 pairs of PC and adjacent non-carcinoma tissues from humans in a tissue microarray (Shanghai Outdo Biotech Co. Ltd, OD-CT-DgPan03-002, 19 male and 11 female) were used for immunohistochemisical staining. All of the samples were formalin-fixed and embedded in paraffin. The sample sections were deparaffinized and successively rehydrated in xylene and alcohol and washed three times in phosphate buffered saline (PBS). The endogenous peroxidase activity was blocked with 3% hydrogen peroxide in PBS for 10 min, and the sections were then incubated with 0.05% trypsin for 30 minat 37°C. After three washes with PBS, nonspecific binding was blocked with 10% BSA in PBS for 30 min at 37°C. The sections were then incubated overnight at 4°C with an enolase 1 antibody at a dilution of 1:400, and sections incubated with PBS instead of the enolase 1 antibody, served as control. DAB (1:50) was used to detect the enolase 1 with the deposition of a brown reaction product in the nuclei and cytoplasm of the positive cells. The proportion of positively stained cells in each section was averaged from three high-magnification images.
Differences between groups were analyzed by two-tailed Chi-square tests and two-tailed Fisher’s exact tests. The significance level was defined as P < 0.05.
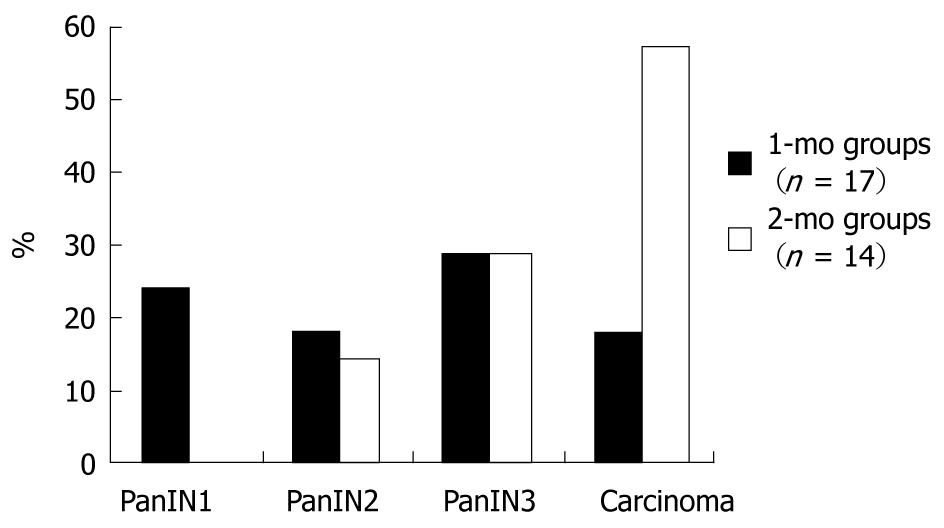
In the 1-mo group, 15% (3/20) of the rats died in the postoperative period, whereas 30% (6/20) died in the 2-mo group. No statistically significant difference in the death rate was observed between the groups (P = 0.4489). Pathologic evaluation revealed that 71% (12/17) of the rats had PanIN lesions and 18% (3/17) of the rats had adenocarcinomas in the 1-mo group. Of these, 24% (4/17) had PanIN1 lesions, 18% (3/17) had PanIN2 lesions and 29% (5/17) had PanIN3 lesions. In the 2-mo group, 14% (2/14) had PanIN2 lesions, 29% (4/14) had PanIN3 lesions, and 57% (8/14) had adenocarcinomas (Figure 1). The difference in the proportions of neoplastic changes was statistically significant between the two groups (P = 0.0488), and there was an increase in the frequency of adenocarcinomas in the 2-mo group compared with the 1-mo group (P = 0.0309). No neoplastic changes were observed in any of the animals in the normal group.
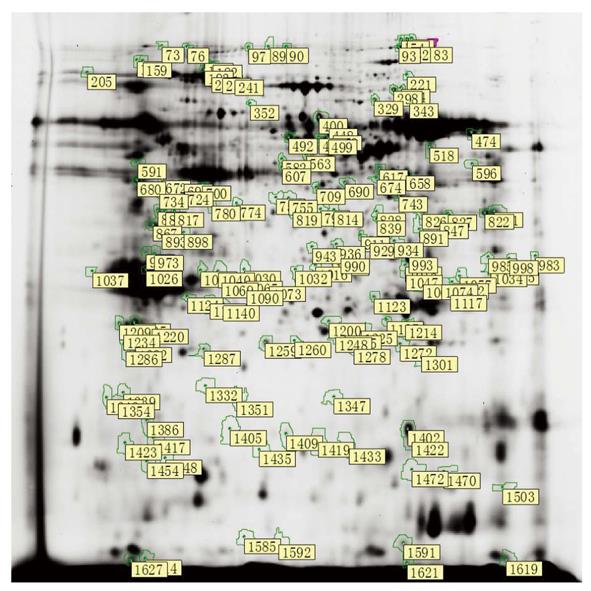
After spot quantification and statistical analysis using the 2D-DIGE approach described above, 1445, 1469, 1380, 1512, and 1637 spots were identified in gel1, gel2, gel3, gel4 and gel5, respectively. After background subtraction and radiometric normalization, matched spots from all of the gels were used for statistical analysis. By using the criterion of P < 0.01, 155 spots were significantly differentially expressed by their relative abundances in groups A, B and C (Figure 2). Furthermore, 31 protein levels progressively increased from normal pancreas to pancreas with DMBA-induced PanIN and PC. Additional 17 protein levels progressively decreased, and 21 spots were selected for identification using a MALDI-TOF/TOF mass spectrometer. We found that the abundances of enolase 1, pancreatic elastase 3B (ELA3B), necdin, Hbp23, chromodomain helicase DNA-binding protein 3 (CHD3), heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1), retinoid X receptor-interacting protein (Rap80), guanine nucleotide-binding protein (G protein), and beta polypeptide 2-like 1 (Gnb2l1) progressively increased with the severity of the disease. The abundances of carboxyl ester lipase (CEL), tumor protein translationally controlled 1 (TPT1), expressed in non-metastatic cells 2 (NME2), phosphoenolpyruvate carboxykinase 2 (PCK2), an unnamed protein product, and glycine C-acetyltransferase progressively decreased with DMBA-induced disease severity (Table 2).
| Spot position | Protein number in NCBI | Protein name | MW | PI | Mascot PMF score | P |
| 230 | Mixture,gi|6753406 + gi|74205924 | Carboxyl ester lipase+unnamed protein product | 70 | 6 | 136 | 4.10E-5 |
| 780 | gi|158186649 | Enolase 1 | 40 | 5.5 | 76 | 5.40E-5 |
| 1205 | gi|6678437 | Tumor protein translationally controlled 1 | 25 | 5 | 98 | 7.90E-5 |
| 1402 | gi|55926145 | Expressed in non-metastatic cells 2 | 20 | 8 | 185 | 0.000 |
| 1132 | gi|149024340 | Pancreatic elastase 3B | 25 | 5.5 | 91 | 0.000 |
| 1066 | gi|56676354 | Necdin | 30 | 8 | 63 | 0.000 |
| 1272 | gi|6435547 | Hbp23 | 25 | 7.5 | 62 | 0.001 |
| 226 | gi|6753406 | Carboxyl ester lipase | 90 | 5.5 | 118 | 0.001 |
| 1405 | gi|109488364 | Chromodomain helicase DNA-binding protein 3 | 20 | 5.5 | 72 | 0.001 |
| 755 | Mixture,gi|149033753 + gi|149039895 | Albumin+retinoid X receptor-interacting protein | 40 | 6.5 | 181 | 0.001 |
| 1259 | gi|12832572 | Unnamed protein product | 25 | 6 | 38 | 0.001 |
| 298 | gi|149063967 | Phosphoenolpyruvate carboxykinase 2 | 66 | 7.5 | 229 | 0.002 |
| 1057 | gi|4504447 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 30 | 8.5 | 62 | 0.002 |
| 993 | gi|5174447 | Guanine nucleotide binding-protein (G-protein), beta polypeptide 2-like 1 | 30 | 7.5 | 146 | 0.003 |
| 1040 | gi|5174447 | Guanine nucleotide binding-protein (G-protein), beta polypeptide 2-like 1 | 30 | 5.5 | 142 | 0.004 |
| 658 | gi|66730435 | Glycine C-acetyltransferase | 40 | 8 | 211 | 0.008 |
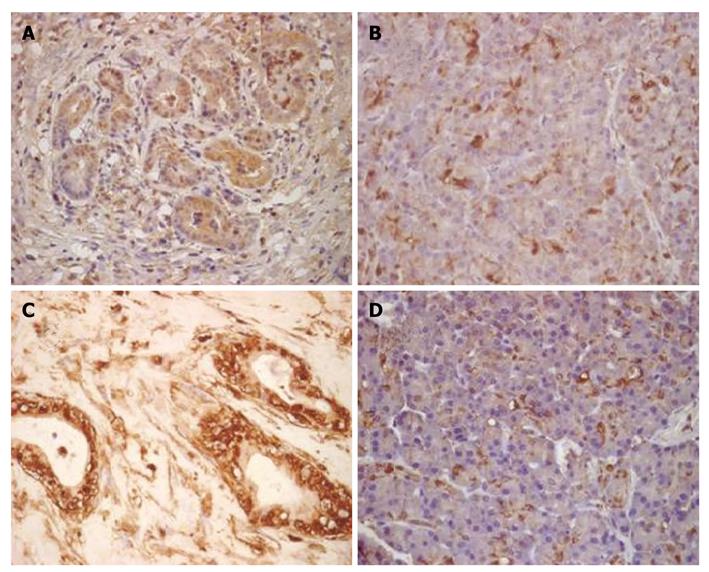
Immunohistochemical staining showed that the expression level of enolase 1 was highest in the nucleus and cytoplasm; membranous immunoreactivity was also detected. The enolase 1 expression levels in DMBA-induced PC and normal rat pancreatic tissues were compared, and the staining results showed that the rat PC tissue had a higher enolase 1 expression level (P = 7.5633E-014). In the rat PC tissue, an average of 83% of cells expressed enolase 1, whereas only 31% of the cells in rat normal pancreatic tissues expressed enolase 1. The expression of enolase 1 was compared in pancreatic cancer and adjacent non-carcinoma tissues from 30 patients, and the average proportion of enolase 1-positive cells in the human pancreatic cancer tissue was 91%, and only 37% in adjacent non-carcinoma tissues. The expression of enolase 1 was also significantly increased in human PC tissues (P = 3.3514E-015) (Figure 3).
Animal models of PanIN and PC that resemble the human disease are very important for research. The characteristics of these models should include the following: (1) A pancreatic ductal phenotype that arises from pancreatic ductal cells; (2) Extensive reactive proliferation of the connective tissue; (3) Obstruction of the bile duct and stomach during disease progression; (4) Early neural, peritoneal, and liver metastases; and (5) High rates of K-ras, P16 and P53 gene mutations[16]. The experimental models of PanIN and PC in the present study are chemical carcinogenesis models. DMBA-induced pancreatic carcinogenesis models resembled human PC in several previous studies. Rivera et al[6] found that DMBA was one of the three different carcinogens reliably producing PC histology that is similar to human ductal adenocarcinoma. DMBA was either implanted directly into the pancreas or infused into the pancreatic ducts of SD rats, and the development of ductal hyperplastic, atypical, and dysplastic changes preceding and accompanying the invasive pancreatic ductal adenocarcinoma could be observed in this experimental model. Jimenez et al[7] demonstrated that DMBA-induced rat tubular complexes and pancreatic adenocarcinomas strongly expressed ductal cell markers (keratin, cytokeratins 19 and 20) but not acinar cell markers (chymotrypsin), suggesting that these tumors arose from ductal cell transformation. K-ras mutations were significantly more frequent in DMBA-induced PC tissues than in normal tissues, with a prevalence of up to 91%[8]. Pancreatic carcinogenesis induced by DMBA implantation in mice was re-evaluated according to the PanIN classification system after its establishment, and extensive pathological changes characteristic of PanIN could be observed one month after carcinogen implantation[9]. We used the PanIN classification system in the histological analysis, and our pathologic evaluation showed that 71% (12/17) of the rats had PanIN lesions and 18% (3/17) had adenocarcinomas in the 1-mo group. In the 2-mo group, 43% (6/14) had PanIN lesions and 57% (8/14) had adenocarcinomas. The frequencies of PanIN and PC in our rat models were similar to a previous study in mice[9]. Our models were satisfactory because the development of precursor lesions (PanIN) and an invasive adenocarcinoma were observed in all groups. There was an increase in the frequency of adenocarcinomas, and the PanIN classification increased with DMBA induction, reflecting the dynamic development of a normal pancreas into a cancerous pancreas.
There are several techniques available for protein separation, both gel- and non-gel-based. Gel-based techniques include traditional two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and, more recently, 2D-DIGE. 2D-DIGE usually contains three samples labeled with three distinct fluorescent dyes: Cy2, Cy3 and Cy5. The Cy2 dye is typically used to label an internal standard; the strength of the internal standard helps map the spots/proteins between the gels, thus rendering different gels more comparable[17-19]. In the current study, we used 2D-DIGE and MALDI-TOF/TOF tandem mass spectrometry to profile pancreatic proteins in rat models of DMBA-induced PanIN and PC and compared these profiles with those of normal rats. Our 2D-DIGE data showed that 155 spots were significantly differentially expressed based on their relative abundances in the three groups. Of these, 31 protein levels progressively increased from normal to PanIN and then to PC tissue, whereas 17 protein levels progressively decreased. These results demonstrated synchronous dynamic changes in the pancreatic protein expression profile after DMBA induction. A number of novel proteins were identified that may be involved in important aspects of mRNA transcription, DNA damage repair, chromatin remodeling, oxidative stress, regulation of tumor growth and metastasis, glucose metabolism, and the synthesis and secretion of pancreatic enzymes.
Enolase 1, also known as α-enolase or non-neuronal enolase (NNE), is an isoenzyme of enolase, which is a glycolytic enzyme catalyzing the conversion of 2-phosphoglycerate into phosphoenolpyruvate. Recent researches have shown that enolase 1 plays an important role in several biological and pathophysiological processes. Enolase 1 is thought to play a potential role in tumorigenesis, cancer invasion and metastasis. Previous proteomic studies reported that enolase 1 was up-regulated in several cancers, such as hepatocellular carcinoma[20-22], non-small lung cancer[23,24], esophageal adenocarcinoma[25], prostate cancer[26], colon cancer[27], oral epithelial and squamous cell carcinoma[28]. In our study, we found a significant up-regulation of enolase 1 in rat models of DMBA-induced PanIN and PC with further verification by the immunohistochemical analysis of human and rat tissues. These results agree with other proteomic researches on human PC. Mikuriya et al[29] found that the expression levels of glycolytic enzymes, including enolase 1, increased in the cancerous pancreatic tissues of 10 patients compared with the paired non-cancerous tissues, as determined by proteomic profiling using two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Shen et al[30] used a proteomic approach using two-dimensional gel electrophoresis and mass spectrometry to identify differentially expressed proteins in six PC cases, two normal adjacent tissues, seven cases of pancreatitis, and six normal pancreatic tissues. Alpha-enolase was also specifically overexpressed in tumors compared with normal and pancreatic tissues.
The hnRNP A2/B1 protein was shown to be up-regulated in our models. This protein plays an important role in the biogenesis and transport of mRNA. The over-expression of hnRNP A2/B1 indicates that normal transcriptional regulation is altered. A previous study also found high levels of hnRNP A2/B1 expression in a limited number of human pancreatic adenocarcinomas from smokers and two pancreatic tumor cell lines, HPAF-11 and SU 86.86[31]. CEL is one kind of pancreatic exocrine enzyme, and we found that it was down-regulated after DMBA induction. Reuss et al[32] verified strong CEL gene expression in the acinar cells of the normal pancreas, and adenocarcinomas showed no expression, which agrees with our results. The other differentially expressed proteins that we identified are rarely reported in PC, and their relationships with PC await further clarification.
In summary, our data have shown DMBA implantation into the pancreas is an effective method to establish rat models of PanIN and PC, and the protein changes observed in DMBA-induced PanIN and PC, such as enolase 1 up-regulation, demonstrate the feasibility of identifying potential molecular targets for the early diagnosis and treatment of PC.
The early detection of pancreatic cancer (PC) is still difficult, but proteomic techniques are emerging as important tools to find potential biomarkers and new therapeutic targets for PC and precancerous pancreatic lesions.
7,12-dimethylbenzanthracene (DMBA) implantation into the pancreas is known to induce PC and pancreatic ductal intraepithelial neoplasia (PanIN) in mice and rats that resemble human PC with K-ras gene mutations and the activation of Notch signaling.
Precancerous pancreas lesions and the early stages of PC among clinical diagnoses are rarely found. In the present research, the authors studied the proteomic variabilities of rat models of DMBA-induced PanIN and PC.
The protein changes in DMBA-induced PanIN and PC reported in this article, such as enolase 1 up-regulation, demonstrate the feasibility of identifying potential molecular targets for early PC diagnostics and therapeutics.
Two-dimensional fluorescence difference gel electrophoresis is a gel-based technique that has recently been used for protein separation. It contains three samples labeled with three distinct fluorescent dyes: Cy2, Cy3 and Cy5.
This is a good study to define the stages of pancreatic carcinoma in experimental models and its correlation with various markers.
Peer reviewer: Dr. Ashok Kumar, MD, Department of Surgical Gastroenterology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, 226014, India
S- Editor Sun H L- Editor Ma JY E- Editor Ma WH
| 1. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-249. [Cited in This Article: ] |
| 2. | Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Klöppel G, Longnecker DS. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579-586. [Cited in This Article: ] |
| 3. | Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, Biankin SA, Compton C, Fukushima N, Furukawa T. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977-987. [Cited in This Article: ] |
| 4. | Dissin J, Mills LR, Mains DL, Black O Jr, Webster PD 3rd. Experimental induction of pancreatic adenocarcinoma in rats. J Natl Cancer Inst. 1975;55:857-864. [Cited in This Article: ] |
| 5. | Bockman DE. Cells of origin of pancreatic cancer: experimental animal tumors related to human pancreas. Cancer. 1981;47:1528-1534. [Cited in This Article: ] |
| 6. | Rivera JA, Graeme-Cook F, Werner J, Z’graggen K, Rustgi AK, Rattner DW, Warshaw AL, Fernández-del Castillo C. A rat model of pancreatic ductal adenocarcinoma: targeting chemical carcinogens. Surgery. 1997;122:82-90. [Cited in This Article: ] |
| 7. | Jimenez RE, Z’graggen K, Hartwig W, Graeme-Cook F, Warshaw AL, Fernandez-del Castillo C. Immunohistochemical characterization of pancreatic tumors induced by dimethylbenzanthracene in rats. Am J Pathol. 1999;154:1223-1229. [Cited in This Article: ] |
| 8. | Z’graggen K, Warshaw AL, Werner J, Graeme-Cook F, Jimenez RE, Fernández-Del Castillo C. Promoting effect of a high-fat/high-protein diet in DMBA-induced ductal pancreatic cancer in rats. Ann Surg. 2001;233:688-695. [Cited in This Article: ] |
| 9. | Osvaldt AB, Wendt LR, Bersch VP, Backes AN, de Cássia A Schumacher R, Edelweiss MI, Rohde L. Pancreatic intraepithelial neoplasia and ductal adenocarcinoma induced by DMBA in mice. Surgery. 2006;140:803-809. [Cited in This Article: ] |
| 10. | Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T. Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci. 2007;98:155-162. [Cited in This Article: ] |
| 11. | Wendt LR, Osvaldt AB, Bersch VP, Schumacher Rde C, Edelweiss MI, Rohde L. Pancreatic intraepithelial neoplasia and ductal adenocarcinoma induced by DMBA in mice: effects of alcohol and caffeine. Acta Cir Bras. 2007;22:202-209. [Cited in This Article: ] |
| 12. | Bersch VP, Osvaldt AB, Edelweiss MI, Schumacher Rde C, Wendt LR, Abreu LP, Blom CB, Abreu GP, Costa L, Piccinini P. Effect of nicotine and cigarette smoke on an experimental model of intraepithelial lesions and pancreatic adenocarcinoma induced by 7,12-dimethylbenzanthracene in mice. Pancreas. 2009;38:65-70. [Cited in This Article: ] |
| 13. | Boros LG, Lerner MR, Morgan DL, Taylor SL, Smith BJ, Postier RG, Brackett DJ. [1,2-13C2]-D-glucose profiles of the serum, liver, pancreas, and DMBA-induced pancreatic tumors of rats. Pancreas. 2005;31:337-343. [Cited in This Article: ] |
| 14. | Rajcevic U, Niclou SP, Jimenez CR. Proteomics strategies for target identification and biomarker discovery in cancer. Front Biosci. 2009;14:3292-3303. [Cited in This Article: ] |
| 15. | Latterich M, Abramovitz M, Leyland-Jones B. Proteomics: new technologies and clinical applications. Eur J Cancer. 2008;44:2737-2741. [Cited in This Article: ] |
| 16. | Ottenhof NA, Milne AN, Morsink FH, Drillenburg P, Ten Kate FJ, Maitra A, Offerhaus GJ. Pancreatic intraepithelial neoplasia and pancreatic tumorigenesis: of mice and men. Arch Pathol Lab Med. 2009;133:375-381. [Cited in This Article: ] |
| 17. | Kondo T. Tissue proteomics for cancer biomarker development: laser microdissection and 2D-DIGE. BMB Rep. 2008;41:626-634. [Cited in This Article: ] |
| 18. | Westermeier R, Scheibe B. Difference gel electrophoresis based on lys/cys tagging. Methods Mol Biol. 2008;424:73-85. [Cited in This Article: ] |
| 19. | Minden J. Comparative proteomics and difference gel electrophoresis. Biotechniques. 2007;43:739, 741, 743 passim. [Cited in This Article: ] |
| 20. | Kuramitsu Y, Nakamura K. Current progress in proteomic study of hepatitis C virus-related human hepatocellular carcinoma. Expert Rev Proteomics. 2005;2:589-601. [Cited in This Article: ] |
| 21. | Takashima M, Kuramitsu Y, Yokoyama Y, Iizuka N, Fujimoto M, Nishisaka T, Okita K, Oka M, Nakamura K. Overexpression of alpha enolase in hepatitis C virus-related hepatocellular carcinoma: association with tumor progression as determined by proteomic analysis. Proteomics. 2005;5:1686-1692. [Cited in This Article: ] |
| 22. | Hamaguchi T, Iizuka N, Tsunedomi R, Hamamoto Y, Miyamoto T, Iida M, Tokuhisa Y, Sakamoto K, Takashima M, Tamesa T. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008;33:725-731. [Cited in This Article: ] |
| 23. | Chang GC, Liu KJ, Hsieh CL, Hu TS, Charoenfuprasert S, Liu HK, Luh KT, Hsu LH, Wu CW, Ting CC. Identification of alpha-enolase as an autoantigen in lung cancer: its overexpression is associated with clinical outcomes. Clin Cancer Res. 2006;12:5746-5754. [Cited in This Article: ] |
| 24. | He P, Naka T, Serada S, Fujimoto M, Tanaka T, Hashimoto S, Shima Y, Yamadori T, Suzuki H, Hirashima T. Proteomics-based identification of alpha-enolase as a tumor antigen in non-small lung cancer. Cancer Sci. 2007;98:1234-1240. [Cited in This Article: ] |
| 25. | Zhao J, Chang AC, Li C, Shedden KA, Thomas DG, Misek DE, Manoharan AP, Giordano TJ, Beer DG, Lubman DM. Comparative proteomics analysis of Barrett metaplasia and esophageal adenocarcinoma using two-dimensional liquid mass mapping. Mol Cell Proteomics. 2007;6:987-999. [Cited in This Article: ] |
| 26. | van den Bemd GJ, Krijgsveld J, Luider TM, van Rijswijk AL, Demmers JA, Jenster G. Mass spectrometric identification of human prostate cancer-derived proteins in serum of xenograft-bearing mice. Mol Cell Proteomics. 2006;5:1830-1839. [Cited in This Article: ] |
| 27. | Katayama M, Nakano H, Ishiuchi A, Wu W, Oshima R, Sakurai J, Nishikawa H, Yamaguchi S, Otsubo T. Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg Today. 2006;36:1085-1093. [Cited in This Article: ] |
| 28. | Ito S, Honma T, Ishida K, Wada N, Sasaoka S, Hosoda M, Nohno T. Differential expression of the human alpha-enolase gene in oral epithelium and squamous cell carcinoma. Cancer Sci. 2007;98:499-505. [Cited in This Article: ] |
| 29. | Mikuriya K, Kuramitsu Y, Ryozawa S, Fujimoto M, Mori S, Oka M, Hamano K, Okita K, Sakaida I, Nakamura K. Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int J Oncol. 2007;30:849-855. [Cited in This Article: ] |
| 30. | Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018-9026. [Cited in This Article: ] |
| 31. | Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183:215-220. [Cited in This Article: ] |
| 32. | Reuss R, Aberle S, Klingel K, Sauter M, Greschniok A, Franke FE, Padberg W, Blin N. The expression of the carboxyl ester lipase gene in pancreas and pancreatic adenocarcinomas. Int J Oncol. 2006;29:649-654. [Cited in This Article: ] |