
Gastrointestinal cancers: current biomarkers in esophageal and gastric adenocarcinoma
Introduction
Esophageal and gastric adenocarcinomas are estimated to represent over 45,000 cases in the USA in 2019 and rank as the 15th (stomach) and 18th (esophageal) most common malignancies in the United States (1). Many cases are diagnosed at an advanced stage and as such, have a dismal prognosis. Early stage carcinomas are typically treated with multimodality therapy, which results in improved survival rates compared to surgery alone. Despite aggressive treatments of potentially curative esophageal cancer, about 50% of patients ultimately developed recurrent disease. Cure in the setting of locally advanced or metastatic disease is exceedingly rare. Systemic therapy is the cornerstone of treatment in the advanced setting. A number of biomarkers that can guide treatment options for patients with esophagogastric adenocarcinoma (EGA) have been identified and these biomarkers will be the focus of this review.
Human epidermal growth factor receptor 2 (HER2/ERBB2)
Human epidermal growth factor receptor (HER) represents a family of receptors, of which HER2 is a member. These proteins are a group of tyrosine kinase receptors that are essential in regulating cellular proliferation, differentiation and survival, but also play a pivotal role in the pathogenesis of several human cancers. The family includes HER1 (ErbB1), HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4) (2). All four receptors have common structural features, which include a extracellular ligand-binding domain that is rich in cysteine residues, a lipophilic transmembrane domain and tyrosine kinase catalytic activity present within the intracellular protein domain (3). HER receptors are located on the cell surface as monomers and undergo dimerization and transphosphorylation of intracellular domains when ligand binds to the extracellular domain. HER2 is an exception in that it does not have a ligand. Activation of HER2-mediated signaling occurs by either by homodimerization (when in high concentrations) or heterodimerization with ligand-activated EGFR or HER3. Dimerization of the receptors results in the tyrosine residues within the cytoplasmic domain to become autophosphorylated with the activation of downstream pathways including protein kinase C (PKC), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), Janus kinase/signal transducer and activator of transcription (JAK/STAT), and mitogen-activated protein kinase (MAPK) which leads to cell proliferation, differentiation, invasion, and angiogenesis (4,5). HER2 containing heterodimers have the greatest mitogenic potential among the HER complexes. The heterodimer pair of HER2-HER3 in particular is the most powerful stimulator of the PI3K signaling cascade, which regulates cell growth and survival (6,7).
Abnormal HER2 signaling has been identified in a number of different cancers, and overexpression and/or amplification of HER2 is a poor prognostic marker in several malignancies (8). In the context of esophageal and gastric adenocarcinoma, prognostic data are conflicting, and large studies fail to show correlation with overall survival (OS) (9-13).
Overexpression
HER2 overexpression is seen in up to ~20% of esophageal and gastric cancers (14,15).. Moreover, overexpression in the stomach varies with histologic subtype, with HER2 being more frequently noted in intestinal-type carcinomas compared to diffuse-type (or poorly cohesive) carcinomas and in tumors that are well to moderately-differentiated compared to those that are poorly-differentiated (15-17). In contrast to breast cancer, the expression of HER2 in esophageal and gastric cancers is quite heterogeneous (13,18).
Testing
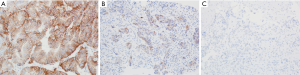
Immunohistochemistry (IHC) and in situ hybridization (ISH) techniques are appropriate methods to assess HER2 overexpression according to the guidelines outlined by the College of American Pathologists (CAP), American Society of Clinical Oncology (ASCO) and the American Society for Clinical Pathology (ASCP) (19). Resection and biopsy specimens from patients with esophageal or gastric tumors should be placed in 10% neutral buffered formalin within 1 hours of collection (cold ischemia time) and allowed to fix to a minimum of 6 and a maximum of 72 hours. IHC should be used as the first testing modality to assessment HER2 expression. HER2 IHC evaluates membranous protein expression in the cancer cells. A four tiered scoring system for HER2 was developed by Hofmann et al., using an assessment area containing at least 10% expression within tumor cells in a resection specimen while a single small cluster of cells (containing at least 5 neoplastic cells) was required for biopsy samples (20). The “completeness” or full membranous staining that is a requirement in HER2 assessment in breast cancers is not seen in gastric or esophageal adenocarcinomas; these tumors typically have a basolateral staining pattern. Therefore, intensity and percentage of HER2 expression on the tumor cells is assessed using a scale from 0 to 3+ (see Table 1). HER2-positive esophageal and gastric cancer is defined as tumors with an IHC score of 3+ (see Figure 1). Scores of 0 or 1+ are considered HER2 negative. For cases exhibiting a HER2 score of 2+ (i.e., equivocal), additional testing using FISH or other ISH techniques should be performed to confirm HER2 status.

Full table
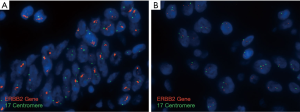
A wide array of ISH techniques can be used for detection of HER2 gene amplification. Some of these are fluorescence in situ hybridization (FISH), chromogenic in situ hybridization (CISH), silver-enhanced in situ hybridization (SISH), and dual in situ hybridization (DISH). While a single HER2(ERBB2) probe may be used for the detection of gene copy number, most assays typically include a chromosome enumeration probe to more accurately calculate the ratio of HER2(ERBB2) signals to copies of chromosome 17 (CEP17) (see Figure 2). A minimum of 20 discrete tumor nuclei are evaluated for the HER2 and CEP17 probe signals. According to the 2016 CAP/ASCP/ASCO guidelines, HER2(ERBB2) amplification is defined as HER2 (ERBB2):CEP17 ratio of ≥2. In cases where the IHC score is 2+ (equivocal), an average of 3 or more CEP17 signals are present with a ratio ≤2, then the presence of more than 6 HER2 signals is interpreted as positive for HER2 amplification by ISH/FISH. In cases where ≤4 HER2 signals are present, the findings are interpreted as negative for HER2 amplification. For cases where a range of 4–6 signals are identified, it is recommended that an additional 20 cells be scored in a separate area or another sample be submitted for evaluation (19).
IHC and FISH have been studied extensively, while next generation sequencing (NGS) for the identification of HER2 amplification is still in its early stages. NGS is often utilized when tissue samples are limited and patients are unable to undergo additional sampling. NGS testing allows for HER2 copy number and HER2 mutations to be assessed at the same time. At present, there are limited data assessing copy number variation (CNV) compared to the current gold-standard methods of IHC and ISH. Unlike in breast cancer, the concordance of HER2 copy number via NGS in gastric cancer was only moderate compared to IHC and ISH (21). The larger discordance in gastric cancer may be a function of underlying biological mechanisms and further studies are warranted. HER2 copy number in liquid biopsies or circulating tumor deoxyribonucleic acid (ctDNA) is being examined for disease burden and capability to predict trastuzumab efficacy in patients with gastric cancer (22). Tumor tissue and ctDNA samples had a high HER2 amplification concordance, signifying that ctDNA may serve as an alternative method for screening HER2-targeted populations, however additional studies are needed.
Targeting therapy
Trastuzumab is a monoclonal antibody that binds to the extracellular domain of the HER2 receptor and prevents the activation of its intracellular tyrosine kinase. In the US, trastuzumab combined with chemotherapy is approved for the 1st line treatment of HER2 amplified esophageal and gastric adenocarcinomas. This approval is based on the positive results from the Trastuzumab for Gastric Cancer (ToGA) trial, which was an open-label, international, phase 3 randomized controlled trial using the anti-HER2 humanized monoclonal antibody trastuzumab. This study demonstrated a median survival of 13.1 months for patients who received trastuzumab and chemotherapy and 11.7 months for patients who were treated with chemotherapy alone (15). Trastuzumab was most effective in HER2-positive tumors. As such, trastuzumab is recommended for tumors with IHC score of 3+ and those with an IHC score of 2+ with evidence of HER2(ERBB2) amplification by ISH. In patients who were found to be positive by ISH, but had IHC scores of 0 or 1+, there was no significant survival benefit seen for these patients and as such trastuzumab is not recommended in this setting (15,23,24).
DNA mismatch repair (MMR)/microsatellite instability (MSI)
DNA MMR is a decidedly preserved mechanism, which exists in organisms from prokaryotes to eukaryotes. MMR is involved in genomic fidelity by repairing DNA after mismatching inaccuracies occur during replication and recombination (25,26). MMR has also been shown to be involved in cell cycle regulation and apoptotic response to a variety of DNA damage (27,28).
Four genes regulate the MMR mechanism: mutL homologue 1 (MLH1), postmeiotic segregation increased 2 (PMS2), and mutS homologue 6 (MSH6). The proteins form heterodimers, namely MLH1/PMS2 and MSH2/MSH6. The MSH2/MSH6 heterodimer is responsible for recognizing base mismatches and insertion-deletion loops, while successive enlistment of the MLH1/PMS2 heterodimer excises the altered stretch of base pairs and resynthesizes the corrected DNA bases in this mismatch site.
The biallelic inactivation of one of the MMR genes can result from mutations (either somatic or germline) or from epigenetic silencing (25,29). When one or more of the MMR proteins are not expressed, the result is referred to as deficient mismatch repair (dMMR); while when all proteins are intact, the status is considered proficient mismatch repair (pMMR). The loss of MLH1 expression leads to inactivation of the MLH1/PMS2 heterodimer and degradation of PMS2, while loss of MSH2 expression leads to inactivation of the MSH2/MSH6 heterodimer and loss of MSH6.
dMMR was identified as a germline mutation in Lynch syndrome patients in 1993 (30). Patients with Lynch syndrome were found to have increased number of microsatellites, or short tandem repeats, in their DNA. Microsatellites are repeating DNA sequences ranging in length from one nucleotide and up to six nucleotides which can be identified within both coding and non-coding regions of the genome (31). These repeating DNA sequences are sensitive to mismatch errors; when MMR systems are deficient (dMMR), there is an accumulation of mutations defined as MSI that can be detected when analyzing polyA microsatellites by polymerase chain reaction. Both MSI and chromosomal instability represent a mutator phenotype; while MSI is a marker of dMMR, not all hypermutated cells will be dMMR/MSI-H (32).
MSI can be seen in up to 5% of esophageal adenocarcinoma. Similar to its colorectal counterpart, MSI-H esophageal adenocarcinomas are histologically distinct in that they have increased tumor infiltrating lymphocytes and high-grade histology, such as medullary, mucinous or signet-ring cell subtype (33). MSI in gastric adenocarcinomas is noted in about 6–9% of cases and is observed in older females, tumors located in the antrum, tumors with intestinal subtype, early stage and are associated with a better prognosis (34-36). Lynch syndrome associated esophageal and gastric adenocarcinoma is rare. The frequency of gastric cancer in Lynch syndrome patients is estimated to be ~1.6% (37), while esophageal adenocarcinoma has not been associated with Lynch syndrome. It should be noted that although 3–5% of esophageal carcinoma cases appears to have MMR deficiency, this is due to somatic and not germline mutations (38).
Testing
Testing to assess the functionality of the MMR system is becoming increasingly common as it has important implications for the screening of Lynch syndrome. MMR status is also prognostic and predictive of treatment response to immune checkpoint inhibitors. Therefore, MMR deficiency should be assessed in patients with locally advanced, recurrent or metastatic esophageal or gastric adenocarcinoma who are candidates for treatment with PD-1 inhibitors. Two clinically utilized testing platforms are used to detect a deficient MMR system in cancer. MSI testing by polymerase chain reaction (PCR) is used to detect instability in microsatellite repeats, while IHC is used to detect the presence or absence of nuclear expression of one or more of the MMR proteins.
IHC
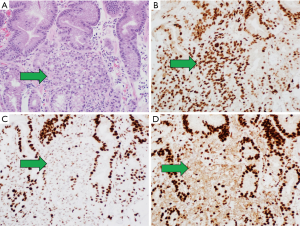
There are commercially available antibodies directed against the MMR proteins MLH1, MSH2, MSH6, and PMS2. IHC detection systems are used to identify the presence or absence of nuclear protein expression. Testing can be performed on formalin-fixed, paraffin-embedded (FFPE) tumor tissue from a biopsy or the resected surgical specimen (see Figure 3). Loss of nuclear expression in one or more the MMR proteins has good correlation with DNA-based MSI testing (39). IHC is widely available, routinely used, and results reported within 48 hours in most pathology laboratories.
MSI testing by PCR
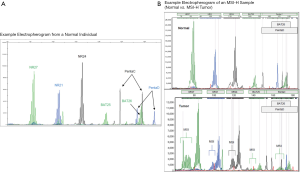
Molecular analysis can be conducted on DNA extracted from fresh, frozen, or FFPE tumor tissue using a PCR-based assay for the detection of MSI. In order to minimize the variability between different MSI tests, the National Cancer Institute (NCI) in 1997 recommended a panel of five microsatellite markers for testing termed the NCI/Bethesda panel. This panel consisted of two mononucleotides loci [big adenine tract (BAT) 25 and BAT26] along with three dinucleotide repeats (D2S123, D5S346 and D17S250) (40). A 2004 NCI workshop recognized the limitations of the original Bethesda panel and recommended the use of a panel of five microsatellite markers, including mononucleotide repeats (BAT25, BAT26, NR21, NR22, and NR 24 or NR27) and dinucleotide repeats (D2S123, D5S346 and D17S250) (Umar 2004, Umar 2004). The NCI panel also provided guidelines for interpretation of MSI-high (MSI-H), MSI-low (MSI-L), and microsatellite stable (MSS) (40). Tumors are classified as MSI-H when there is a documented shift in at least 2 of five tumor loci when compared to non-neoplastic tissue (see Figure 4). If a larger panel is used, then a shift of >30% of loci is required for MSI-H. When there is a shift in one locus (<30% of loci in larger panels), the tumor is classified as MSI-L. When there is no instability detected (<10% loci in larger panels), the tumor is considered MSS. Notably, subsequent studies discovered that a panel of mononucleotide markers, which are often more unstable than dinucleotide markers, was more sensitive than the original NCI panel (41-44).
Targeting therapy
In 2017, the FDA approved pembrolizumab for patients with dMMR/MSI-H solid tumors that are deemed unresectable or metastatic, irrespective of tumor type or site. These patients have progressed on at least one line of prior therapy. This approval is notable as it is the first biomarker-based, disease agnostic approval in oncology. The FDA’s approval of pembrolizumab was based on data from five multicenter, single cohort clinical trials, which enrolled a total of 149 patients with dMMR/MSI-H tumors. Objective responses were seen in 39.6% of subjects, and lasted for 6 months in 78% of the responders. The overall response rate was similar across different tumor types (45).
Programmed death-ligand 1 (PD-L1)
The immune system plays an important role in homeostasis. T-lymphocytes can selectively identify and kill pathogens and tumor cells by coordinating responses by the innate and adaptive immune systems. There are numerous checks and balances so that the immune system does not mistakenly destroy healthy cells during their response. Cancer cells frequently exploit these immune checkpoints in order to evade detection.
Some of these immune checkpoint proteins include programmed cell death protein 1 (PD-1) and PD-L1 that act as co-inhibitory factors that can stop or minimize T-cell responses. PD-1 was originally cloned in 1992 and was felt to be responsible for the process of apoptosis (46). PD-1 is a type I transmembrane protein located on chromosome 2q37, coded by the PDCD1 gene. PD-1 is known to be present on the surface of B- and T-lymphocytes, macrophages and some dendritic cells (47,48). PD-L1 is encoded by the CD274 gene located on chromosome 9p24.2 that results in a 40-kDa transmembrane protein expressed on macrophages and dendritic cells that binds to PD-1 (49). PD-L1 is known to be expressed in a wide variety of non-inflammatory cells including pneumocytes, hepatocytes, endothelial cells, etc. (48,50,51). Moreover, PD-L1 has been shown to have high expression levels in several tumor types including breast carcinoma, hepatocellular carcinoma, squamous cell carcinomas of the head and neck, and lung adenocarcinomas (52-57). Several studies, including those for esophageal and gastric adenocarcinoma, have identified PD-L1 overexpression as a predictive biomarker for response to immune checkpoint inhibitors (55,58).
Testing
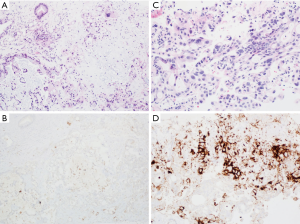
There are commercially available antibodies directed against various clones of PD-L1 that can detect the presence of membranous protein expression. PD-L1 expression is seen in about 40% of gastroesophageal adenocarcinomas (59,60). PD-L1 expression on tumor cell themselves is infrequent, and most of PD-L1 staining is seen in immune cells within the stroma of the tumor, specifically lymphocytes and macrophages (60). As such, results from PD-L1 testing in upper gastrointestinal cancers is reported as combined positive score (CPS) to account for all the cell types that stain positively, in contrast to total positive score (TPS), which is used to analyze lung cancer specimens.
Prior to immunostaining, it should be confirmed that at least 100 viable tumor cells are present within the sample. Immunohistochemistry slides are examined at low magnification to assess the extent of membrane staining of cells within the tumor. At higher magnification, the number of PD-L1 positive cells (tumor cells, lymphocytes and macrophages) relative to total number of viable tumor cells is determined. It should be noted that necrotic debris and stromal cells within the tumor should not be included within the calculation. The results are reported in the setting of esophageal and gastric carcinomas as CPS utilizing the formula:
CPS = [# positive PD-L1 cells (tumor cells, lymphocytes, macrophages)/# PD-L1 positive and PD-L1 negative tumor cells] ×100%
CPS is reported as a single number with a maximum score of 100. CPS is considered positive with a score ≥1, while CPS ≤1 is considered negative (see Figure 5).
Targeting therapy
Pembrolizumab is an anti PD-1 antibody that was approved for advanced esophageal, gastric and GEJ adenocarcinoma in 2017. This approval was based on the results from multi-cohort open label nonrandomized trial (KEYNOTE-059), which enrolled 259 patients who have previously received 2 prior lines of therapy (61). Patients whose tumors were noted to be PD-L1 positive (CPS ≥1) were found to have a response rate of 15.5%, while only 6.4% of patients with PD-L1 negative tumors responded. It was encouraging to see prolonged response duration, ranging between 5.3 to 14.1 months, in this advanced setting. Activity of pembrolizumab was also evaluated in earlier lines of therapy. A phase 3 trial (KEYNOTE-061) for patients with gastric cancer and phase 3 trial (KEYNOTE-181) for patients with esophageal and Siewert type 1 gastroesophageal junction tumors evaluated activity of pembrolizumab in comparison to chemotherapy in the 2nd line setting. These trials did not improve patient OS with immunotherapy treatment. However, responses from pembrolizumab were more durable than those seen with chemotherapy, and responses were more frequent when tumors had higher CPS scores (62-64). In the 1st line setting, KEYNOTE-062, a randomized phase 3 trial examining the role of pembrolizumab alone or in combination with chemotherapy compared to chemotherapy alone failed to meet its primary endpoints (65). Therefore, at the present time, pembrolizumab remains approved for use in PD-L1 positive tumors with CPS ≥1 after progression on at least 2 prior lines of therapy, as well as for dMMR/MSI-H tumors regardless of PD-L1 expression.
Future biomarkers
A number of novel targets in esophageal and gastric cancer are being studied in clinical trials. As was discussed previously, the status of HER2 is frequently tested using FFPE material from the patient’s esophageal or gastric tumor. Due to treatment modalities or upon disease progression, it is known that the HER2 status may change. Evaluation of HER2 status by next generation sequencing repeatedly during the course of disease is essential for evaluation of secondary resistance to trastuzumab. Moreover, information about the level of HER2 amplification, subclones lacking HER2 amplification, deletion of ERBB2 exon 16, and co-mutations in other signaling partners (for example RAS, MET and PI3K) may predict resistance to treatment and may uncover other clinically relevant targets (66).
Gene fusions that involve neurotrophic-tropomyosin receptor kinase (NTRK) genes are known drivers of oncogenesis. These fusions are rare and were observed in only 0.31% of adult tumors (67). In November 2018 and August 2019, accelerated approval was granted by the FDA to larotrectinib and entrectinib, respectively for patients age 12 years and older for NTRK gene fusion solid tumors that do not have a known resistance mutation. NTRK gene fusions are exceedingly rare in gastric and esophageal carcinoma and their response to this targeted therapy needs to be further explored.
Recent studies demonstrate a fusion between CLDN18 and ARHGAP26 genes in ~30% of young patients with gastric cancer. The CLDN18 gene codes for claudin-18 that is responsible for tight junctions in epithelial cells. Rho GTPase activating protein 26 (ARHGAP26) gene codes for a multidomain protein that is necessary for endocytosis. The frequency of this fusion is much higher than most other reported driver mutations and druggable/targetable genes from other types of cancers, supporting its role in tumorigenesis (68-70). Nearly all CLDN18–ARHGAP26/6 fusion positive gastric carcinomas have been shown to express CLDN18.2 protein (71). IMAB362 (zolbetuximab) is a novel chimeric IgG1 antibody highly specific for CLDN18.2 and recent preclinical studies and Phase II clinical trials are showing promising results (72,73).
With recent research examining the molecular classification of gastric cancer into several subtypes, the role of fibroblast growth factor receptor (FGFR) has come to the forefront with up to 9% of gastric cancer patients being identified with FGFR2 amplifications (68,74). Due to the increased toxicity and inability to accurately predict response, the first generation tyrosine kinase inhibitors have fallen out of favor. With the release of second generation, selective pan-FGFR inhibitors, there is a renewed interest in this area and several Phase 1 trials are enrolling FGFR-aberrant tumors.
Additional biomarkers are being explored in an attempt to better define the patient population most likely to respond to immunotherapy treatments. Some of the markers currently under investigation include lymphocyte-activation gene 3 (LAG3), T-cell immunoglobulin and mucin-domain containing-3 (TIM3), and tumor mutational burden.
Conclusions
Although there have been several biomarkers identified that can aid in the selection of treatment modalities, treatment outcomes in advanced disease in esophageal and gastric adenocarcinoma remains poor. Identification of new biomarkers, moving existing biomarkers into earlier lines of therapy, and evaluating new combinations of existing biomarkers are and should be the focus of future research efforts. It is imperative to continue to identify more effective biomarkers to predict the effectiveness of therapy before treatment. It is with great hope that we look to upcoming clinical trials to shed more light on novel biomarkers and targeted therapies.
Acknowledgments
We thank Virginia Kurth for assistance with case identification and sample acquisition that as a result has greatly improved the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Ward E, Sherman RL, Henley SJ, et al. Annual Report to the Nation on the Status of Cancer, 1999-2015, Featuring Cancer in Men and Women ages 20-49. J Natl Cancer Inst 2019;111:1279-97. [Crossref] [PubMed]
- Riese DJ 2nd, Stern DF. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays 1998;20:41-8. [Crossref] [PubMed]
- Brandt-Rauf PW, Pincus MR, Carney WP. The c-erbB-2 protein in oncogenesis: molecular structure to molecular epidemiology. Crit Rev Oncog 1994;5:313-29. [Crossref] [PubMed]
- Yakes FM, Chinratanalab W, Ritter CA, et al. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res 2002;62:4132-41. [PubMed]
- Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007;26:6469-87. [Crossref] [PubMed]
- Neve RM, Lane HA, Hynes NE. The role of overexpressed HER2 in transformation. Ann Oncol 2001;12 Suppl 1:S9-13. [Crossref] [PubMed]
- Menard S, Pupa SM, Campiglio M, et al. Biologic and therapeutic role of HER2 in cancer. Oncogene 2003;22:6570-8. [Crossref] [PubMed]
- Yan M, Schwaederle M, Arguello D, et al. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev 2015;34:157-64. [Crossref] [PubMed]
- Yonemura Y, Ninomiya I, Ohoyama S, et al. Expression of c-erbB-2 oncoprotein in gastric carcinoma. Immunoreactivity for c-erbB-2 protein is an independent indicator of poor short-term prognosis in patients with gastric carcinoma. Cancer 1991;67:2914-8. [Crossref] [PubMed]
- Yonemura Y, Ninomiya I, Yamaguchi A, et al. Evaluation of immunoreactivity for erbB-2 protein as a marker of poor short term prognosis in gastric cancer. Cancer Res 1991;51:1034-8. [PubMed]
- Uchino S, Tsuda H, Maruyama K, et al. Overexpression of c-erbB-2 protein in gastric cancer. Its correlation with long-term survival of patients. Cancer 1993;72:3179-84. [Crossref] [PubMed]
- Nakajima M, Sawada H, Yamada Y, et al. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer 1999;85:1894-902. [Crossref] [PubMed]
- Yoon HH, Shi Q, Sukov WR, et al. Association of HER2/ErbB2 expression and gene amplification with pathologic features and prognosis in esophageal adenocarcinomas. Clin Cancer Res 2012;18:546-54. [Crossref] [PubMed]
- Sakai K, Mori S, Kawamoto T, et al. Expression of epidermal growth factor receptors on normal human gastric epithelia and gastric carcinomas. J Natl Cancer Inst 1986;77:1047-52. [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Tanner M, Hollmen M, Junttila TT, et al. Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Oncol 2005;16:273-8. [Crossref] [PubMed]
- Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 2008;19:1523-9. [Crossref] [PubMed]
- Yoon HH, Shi Q, Sukov WR, et al. Adverse prognostic impact of intratumor heterogeneous HER2 gene amplification in patients with esophageal adenocarcinoma. J Clin Oncol 2012;30:3932-8. [Crossref] [PubMed]
- Bartley AN, Washington MK, Ventura CB, et al. HER2 Testing and Clinical Decision Making in Gastroesophageal Adenocarcinoma: Guideline From the College of American Pathologists, American Society for Clinical Pathology, and American Society of Clinical Oncology. Arch Pathol Lab Med 2016;140:1345-63. [Crossref] [PubMed]
- Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008;52:797-805. [Crossref] [PubMed]
- Ross DS, Zehir A, Cheng DT, et al. Next-Generation Assessment of Human Epidermal Growth Factor Receptor 2 (ERBB2) Amplification Status: Clinical Validation in the Context of a Hybrid Capture-Based, Comprehensive Solid Tumor Genomic Profiling Assay. J Mol Diagn 2017;19:244-54. [Crossref] [PubMed]
- Wang H, Li B, Liu Z, et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur J Cancer 2018;88:92-100. [Crossref] [PubMed]
- Bang YJ. Advances in the management of HER2-positive advanced gastric and gastroesophageal junction cancer. J Clin Gastroenterol 2012;46:637-48. [Crossref] [PubMed]
- Van Cutsem E, Bang YJ, Feng-Yi F, et al. HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 2015;18:476-84. [Crossref] [PubMed]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem 1996;65:101-33. [Crossref] [PubMed]
- Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol 2002;191:28-41. [Crossref] [PubMed]
- Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ 2001;8:1076-92. [Crossref] [PubMed]
- Peters AC, Young LC, Maeda T, et al. Mammalian DNA mismatch repair protects cells from UVB-induced DNA damage by facilitating apoptosis and p53 activation. DNA Repair (Amst) 2003;2:427-35. [Crossref] [PubMed]
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998;95:6870-5. [Crossref] [PubMed]
- Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993;75:1027-38. [Crossref] [PubMed]
- Findeisen P, Kloor M, Merx S, et al. T25 repeat in the 3' untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res 2005;65:8072-8. [Crossref] [PubMed]
- Arzimanoglou II, Gilbert F, Barber HR. Microsatellite instability in human solid tumors. Cancer 1998;82:1808-20. [Crossref] [PubMed]
- Farris AB 3rd, Demicco EG, Le LP, et al. Clinicopathologic and molecular profiles of microsatellite unstable Barrett Esophagus-associated adenocarcinoma. Am J Surg Pathol 2011;35:647-55. [Crossref] [PubMed]
- Smyth EC, Wotherspoon A, Peckitt C, et al. Mismatch Repair Deficiency, Microsatellite Instability, and Survival: An Exploratory Analysis of the Medical Research Council Adjuvant Gastric Infusional Chemotherapy (MAGIC) Trial. JAMA Oncol 2017;3:1197-203. [Crossref] [PubMed]
- Polom K, Marano L, Marrelli D, et al. Meta-analysis of microsatellite instability in relation to clinicopathological characteristics and overall survival in gastric cancer. Br J Surg 2018;105:159-67. [Crossref] [PubMed]
- Choi YY, Kim H, Shin SJ, et al. Microsatellite Instability and Programmed Cell Death-Ligand 1 Expression in Stage II/III Gastric Cancer: Post Hoc Analysis of the CLASSIC Randomized Controlled study. Ann Surg 2019;270:309-16. [Crossref] [PubMed]
- Capelle LG, Van Grieken NC, Lingsma HF, et al. Risk and epidemiological time trends of gastric cancer in Lynch syndrome carriers in the Netherlands. Gastroenterology 2010;138:487-92. [Crossref] [PubMed]
- Falkenback D, Johansson J, Halvarsson B, et al. Defective mismatch-repair as a minor tumorigenic pathway in Barrett esophagus-associated adenocarcinoma. Cancer Genet Cytogenet 2005;157:82-6. [Crossref] [PubMed]
- Baudhuin LM, Burgart LJ, Leontovich O, et al. Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer 2005;4:255-65. [Crossref] [PubMed]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57. [PubMed]
- Perucho M. Correspondence re: C.R. Boland et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res., 58: 5248-5257, 1998. Cancer Res 1999;59:249-56. [PubMed]
- Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002;123:1804-11. [Crossref] [PubMed]
- Bacher JW, Flanagan LA, Smalley RL, et al. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers 2004;20:237-50. [Crossref] [PubMed]
- Murphy KM, Zhang S, Geiger T, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn 2006;8:305-11. [Crossref] [PubMed]
- Lemery S, Keegan P, Pazdur R. First FDA Approval Agnostic of Cancer Site - When a Biomarker Defines the Indication. N Engl J Med 2017;377:1409-12. [Crossref] [PubMed]
- Ishida Y, Agata Y, Shibahara K, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 1992;11:3887-95. [Crossref] [PubMed]
- Nishimura H, Agata Y, Kawasaki A, et al. Developmentally regulated expression of the PD-1 protein on the surface of double-negative (CD4-CD8-) thymocytes. Int Immunol 1996;8:773-80. [Crossref] [PubMed]
- Keir ME, Butte MJ, Freeman GJ, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008;26:677-704. [Crossref] [PubMed]
- Dong H, Zhu G, Tamada K, et al. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999;5:1365-9. [Crossref] [PubMed]
- Yamazaki T, Akiba H, Iwai H, et al. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol 2002;169:5538-45. [Crossref] [PubMed]
- Zhong X, Tumang JR, Gao W, et al. PD-L2 expression extends beyond dendritic cells/macrophages to B1 cells enriched for V(H)11/V(H)12 and phosphatidylcholine binding. Eur J Immunol 2007;37:2405-10. [Crossref] [PubMed]
- Qin T, Zeng YD, Qin G, et al. High PD-L1 expression was associated with poor prognosis in 870 Chinese patients with breast cancer. Oncotarget 2015;6:33972-81. [Crossref] [PubMed]
- Lin YM, Sung WW, Hsieh MJ, et al. High PD-L1 Expression Correlates with Metastasis and Poor Prognosis in Oral Squamous Cell Carcinoma. PLoS One 2015;10:e0142656. [Crossref] [PubMed]
- Aguiar PN Jr, Santoro IL, Tadokoro H, et al. The role of PD-L1 expression as a predictive biomarker in advanced non-small-cell lung cancer: a network meta-analysis. Immunotherapy 2016;8:479-88. [Crossref] [PubMed]
- Chen CL, Pan QZ, Zhao JJ, et al. PD-L1 expression as a predictive biomarker for cytokine-induced killer cell immunotherapy in patients with hepatocellular carcinoma. Oncoimmunology 2016;5:e1176653. [Crossref] [PubMed]
- Zhou Y, Miao J, Wu H, et al. PD-1 and PD-L1 expression in 132 recurrent nasopharyngeal carcinoma: the correlation with anemia and outcomes. Oncotarget 2017;8:51210-23. [PubMed]
- Zhou Y, Shi D, Miao J, et al. PD-L1 predicts poor prognosis for nasopharyngeal carcinoma irrespective of PD-1 and EBV-DNA load. Sci Rep 2017;7:43627. [Crossref] [PubMed]
- Aguiar PN Jr, Santoro IL, Tadokoro H, et al. A pooled analysis of nivolumab for the treatment of advanced non-small-cell lung cancer and the role of PD-L1 as a predictive biomarker. Immunotherapy 2016;8:1011-9. [Crossref] [PubMed]
- Wu C, Zhu Y, Jiang J, et al. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem 2006;108:19-24. [Crossref] [PubMed]
- Thompson ED, Zahurak M, Murphy A, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017;66:794-801. [Crossref] [PubMed]
- Fuchs CS, Doi T, Jang RW, et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol 2018;4:e180013. [Crossref] [PubMed]
- Shitara K, Ozguroglu M, Bang YJ, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet 2018;392:123-33. [Crossref] [PubMed]
- Fuchs CS, Ozguroglu M, Bang YJ, et al. Pembrolizumab (pembro) vs paclitaxel (PTX) for previously treated advanced gastric or gastroesophageal junction (G/GEJ) cancer: Phase 3 KEYNOTE-061 trial. 2018 ASCO Annual Meeting, Oral abstract presentation; June 4, 2018.
- Kojima T, Muro K, Francois E, et al. Pembrolizumab versus chemotherapy as second-line therapy for advanced esophageal cancer: Phase III KEYNOTE-181 study. 2019 ASCO Annual Meeting, Oral abstract presentation; January 17, 2019.
- Tabernero J, Van Cutsem E, Bang YJ, et al. Pembrolizumab with or without chemotherapy versus chemotherapy for advanced gastric or gastroesophageal junction (G/GEJ) adenocarcinoma: The phase III KEYNOTE-062 study. 2019 ASCO Annual Meeting, Oral abstract presentation; June 2, 2019.
- Janjigian YY, Sanchez-Vega F, Jonsson P, et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer Discov 2018;8:49-58. [Crossref] [PubMed]
- Okamura R, Boichard A, Kato S, et al. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis Oncol 2018;2018.
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Shu Y, Zhang W, Hou Q, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun 2018;9:2447. [Crossref] [PubMed]
- Nakayama I, Shinozaki E, Sakata S, et al. Enrichment of CLDN18-ARHGAP fusion gene in gastric cancers in young adults. Cancer Sci 2019;110:1352-63. [Crossref] [PubMed]
- Tanaka A, Ishikawa S, Ushiku T, et al. Frequent CLDN18-ARHGAP fusion in highly metastatic diffuse-type gastric cancer with relatively early onset. Oncotarget 2018;9:29336-50. [Crossref] [PubMed]
- Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol 2017;10:105. [Crossref] [PubMed]
- Sahin U, Schuler M, Richly H, et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur J Cancer 2018;100:17-26. [Crossref] [PubMed]
- Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449-56. [Crossref] [PubMed]
Cite this article as: Dhakras P, Uboha N, Horner V, Reinig E, Matkowskyj KA. Gastrointestinal cancers: current biomarkers in esophageal and gastric adenocarcinoma. Transl Gastroenterol Hepatol 2020;5:55.


