
Beta-1 syntrophin (SNTB1) regulates colorectal cancer progression and stemness via regulation of the Wnt/β-catenin signaling pathway
Introduction
Colorectal cancer (CRC) is a common and lethal malignancy that represents the second leading cause of death by cancer worldwide. The mortality of CRC can be attributed to therapeutic failure and distant metastasis. For potentially resectable CRC, the combination of surgical removal and systemic chemotherapy significantly reduces the mortality rate of CRC patients. However, a small fraction of these patients will inevitably develop a tumor recurrence, generally in the form of metastasis. Recent evidence suggests that not all cell populations that are present in primary cancers are capable of regenerating a tumor as isolated entities (1). The heterogeneity among cancer cell populations with different proliferative, differentiative, and self-renewal potential has led to the development of the “cancer stem cell” theory. Cancer stem cells, or tumor-initiating cells, which can be identified by surface markers such as CD133 (2) and CD44 (3), possess the capacity to propagate a tumor when implanted into immunodeficient mice. There are currently two main approaches to identifying and isolating cancer stem cells. The first approach is flow cytometric isolation by cancer stem cell-specific cell surface marker expressions like CD133, epithelial cell adhesion molecule (EpCAM) (3), or leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5) (4). The second involves isolation by the side population (SP) phenotype that effluxes Hoechst 33342 dye. SPs are an enriched source of cancer stem cells that are characterized by their capacity for Hoechst 33342 exclusion. The existence of SP cells has been reported in a variety of tumors, including ovarian, esophageal, lung, and colorectal carcinoma.
Wnt/β-catenin signaling is one of the well-studied signaling pathways that regulate tumorigenesis (5-7). It has been reported that mutations on the Wnt signaling pathway occur in approximately 94% of CRCs (8). And almost all mutations in the Wnt signaling cascade in CRC eventually cause the accumulation of β-catenin. Consistently, aberrant activation of β-catenin is commonly found in CRC (more than 80%) (9) and is correlated with the poor prognosis of CRC patients (10,11). In the Wnt-β-catenin pathway, upon binding with Wnt ligands, the cytoplasmic β-catenin is prevented from degradation because of the inactivation of the destruction complex. The accumulation of β-catenin in cytosol is then translocated to the nucleus where it forms a complex with members of the lymphoid enhancer binding factor/T-cell specific transcription factor (LEF/TCF) transcriptional factors (12). Subsequently, coactivators like the CREB binding protein (CBP) or p300 are further recruited to this complex, thereby triggering a series of complicated processes that direct cell proliferation and stemness maintenance. c-Myc is one of the molecular targets of β-catenin that modulates a variety of cellular signals, including cell cycle progression and metabolism (13). As an example, cyclin D1 (CCND1), a key component in regulating the cell cycle G1/S checkpoint, is one of the well-studied downstream target genes of β-catenin-c-Myc-modulated transcription (12,14).
The β-catenin pathway is also reported to regulate the maintenance of cancer stem cells and affect their differentiation in CRC (15). β-catenin modulates the homeostatic balance between the self-renewal and differentiation state depending on the coactivator recruited. Specifically, the β-catenin-CBP interaction favors stemness maintenance, while the β-catenin-p300 interaction stimulates differentiation (16). Interestingly, two of the most commonly reported cancer stem cell markers in CRC, EpCAM and CD44, have been characterized as transcriptional targets of the Wnt/β-catenin pathway (17). Moreover, it has been reported that reduction of β-catenin downregulates EpCAM gene expression and that TCF/β-catenin-binding inhibitors suppress EpCAM-positive cell growth in hepatocellular carcinoma (HCC) (18).
Syntrophins are cytoplasmic membrane-associated adaptor proteins that serve to provide a platform for the formation of cellular signal transduction complexes (19). The syntrophin family of proteins consists of five isoforms: the alpha-1 syntrophin (SNTA1), beta-1 syntrophin (SNTB1), beta-2 syntrophin (SNTB2), gamma-1 syntrophin (SNTG1), and gamma-2-syntrophin (SNTG2). Of these, SNTA1 is the most extensively studied and is primarily expressed in skeletal and cardiac muscles (20,21). SNTB1 and SNTB2 are ubiquitously expressed and are present in almost all mammalian tissues, while SNTG1 and SNTG2 are expressed uniquely in the brain. It is believed that the unique tissue distribution pattern and distinct subcellular localization of these syntrophin isoforms may imply their diverse biological functions (22). By direct interaction with dystrophin, syntrophins serve as scaffolding proteins with multiple protein interaction domains that bridge cell surface receptors, ion channels, and downstream effectors to the dystrophin-associated protein complex, thereby modulating and coordinating various intracellular signaling events and crucial cellular processes (22). Although the function of dystrophin has been well studied in muscle and neurons, much less is known about syntrophins (23).
In the present paper, we confirmed the upregulation of SNTB1 in CRC tissues and CRC cell lines by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and Western blot analysis. We also revealed the positive correlation between SNTB1 expression levels and CRC clinical prognostic factors and showed that elevated SNTB1 expression promoted tumorigenesis by increasing the cellular levels of β-catenin. Silencing SNTB1 expression significantly downregulated several β-catenin downstream proteins, such as c-Myc and cell cycle modulator CCND1, and, importantly, decreased the percentage and clonogenicity of SP cells. Interestingly, the β-catenin agonist SKL2001 was able to counteract these phenotypes caused by SNTB1 knockdown and not only stimulated cancer cell growth but restored the level of SP cells. Therefore, our studies provide insights that SNTB1 may be a novel component of the β-catenin signaling cascade, which is important for modulating the proliferative capacity and maintaining the stemness of cancer-initiating cells. SNTB1 may represent a valuable molecular marker for CRC prognosis, although further studies are necessary to determine the biological role of SNTB1 in tumor development and the underlying molecular mechanisms. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2700).
Methods
Antibodies and chemicals
The following antibodies were used: SNTB1 (Abcam, ab242046, 1:2,000), α-tubulin (CST, #3873s, 1:2,000), CCND1 (CST, #55506, 1:1,000), c-Myc (CST, #18583, 1:1,000), β-catenin (CST, #8480, 1:1,000), CD133 (CST, #64326, 1:2,000), CD44 (CST, #37259, 1:2,000), EpCAM (Abcam, ab223582, 1:2,000), KLF4 (Abcam, ab215036, 1:2,000), ALDH1A1 (Abcam, ab134188, 1:2,000), POU5F1 (Abcam, ab230429, 1:2,000), and LGR5 (Abcam, ab273092, 1:2,000). The β-catenin agonist SKL2001 (Sigma, USA) was dissolved in dimethylsulfoxide (DMSO) and stored at –20 °C.
Database and tissue specimens
The SNTB1 mRNA expression data was obtained from The Cancer Genome Atlas (TCGA) database. The TCGA-COAD dataset contains mRNA sequencing data from 459 colon cancer samples and 41 adjacent normal tissues (ANT), and the TCGA-READ dataset contains mRNA sequencing data from 166 rectal cancer (RC) samples and nine ANT. Tissue specimens were collected from the Affiliated Jiangmen Hospital of Sun Yat-sen University (Guangdong, China) and included 10 pairs of fresh CRC tissues and ANT, 25 cases of precancerous lesions, 85 cases of benign lesions, and 278 cases of malignant colorectal lesions. The clinicopathological details are summarized in Tables S1-S4, respectively. Ethical approval and informed consent for the collection of specimens was endorsed by the Institutional Research Ethics Committee of the Affiliated Jiangmen Hospital of Sun Yat-sen University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture, shRNA vector construction, and transfection
The human CRC cell lines were acquired from the cell bank of the Shanghai Chinese Academy of Sciences (China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in a humidified incubator at 37 °C with 5% CO2. For transfection, SNTB1-specific shRNA cells or non-targeting empty vector (shVec.) cells were synthesized by GenePharma (Suzhou, China). The shRNA sequences are listed in Table S5. Cell transfection was performed using Lipofectamine 3000 (Thermo Fisher, USA) as described in our previous studies (24,25).
RT-qPCR and Western blot
Total RNA extraction and reverse transcription were conducted according to the manufacturer’s instructions. Real-time quantitative PCR was performed using Master Mix containing SYBR Green (TaKaRa, Japan), and the relative gene expressions were calculated by the comparative threshold cycle (2–ΔΔCt) method as described in our previous studies (24,25). Primers are listed in Table S6. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the endogenous control. Western blot was performed according to the standard protocol. Briefly, whole cell lysates were extracted by RIPA lysis buffer, and protein concentration was measured using a standard Bradford assay before loading to SDS-PAGE gel. Protein bands were transferred to a PVDF membrane (Millipore, USA), incubated with appropriate primary antibodies and horseradish peroxidase (HRP)-labeled secondary antibodies, and then visualized by the ECL reagent. Protein expression levels were analyzed by Image J and normalized to α-tubulin unless specified otherwise.
Immunohistochemistry (IHC)
The IHC staining of the tissue samples has recently been described (25,26). In brief, formalin-fixed, paraffin-embedded tissues were deparaffinized in xylene, then hydrated through successive graded ethanol solutions. This was followed by incubation with an anti-SNTB1 primary antibody at 4 °C overnight in a humidified chamber and with a secondary antibody at 37 °C for 1 h. Tissue sections were counterstained with hematoxylin at room temperature. Staining was assessed semi-quantitatively under a light microscope at ×400 magnification by two independent observers, blinded to the origin of the tissue. The staining was evaluated according to the scoring criteria described previously (27). SNTB1 expression levels in CRC tissue samples were evaluated by the following staining index (SI) values: SI ≤4 was defined as weak staining or a low expression of SNTB1, and SI ≥6 was defined as strong staining or a high expression of SNTB1. A score of 4 was the median SI score in all tissue samples.
Xenograft assays in nude mice
Eighteen BALB/c nude mice (15–20 g, aged 4–5 weeks, female) were obtained from the Experimental Animal Center of Southern Medical University (Guangzhou, China) and were maintained in a specific pathogen-free (SPF) laboratory with controlled temperature, humidity, and a 12-hour light/dark cycle. Mice were randomly allocated into three groups and then subjected to subcutaneous injection of SW620 cells (5×106) with either shVec. or SNTB1-targeting shRNA (shSNTB1) transduction. The tumor volume was measured by an external caliper. The tumor weight was measured after the mice were sacrificed by cervical dislocation. The excised tumor tissues were stored in liquid nitrogen for subsequent characterization. Animal experiments in the study were approved by the Animal Experimental Ethics Committee of Guangdong Medical University (GDY1902070) in compliance with the National Guidelines for the Care and Use of Animals.
Tumor sphere formation assay
For the tumor sphere formation assay, single-cell suspensions were plated in low-attachment 6-well plates and cultured in the DMEM medium supplemented with B-27, epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF). The cells were then cultivated for 7 to 10 days to form spheres. Afterward, the spheres were counted and photographed under the microscope.
Flow cytometry-based SP analysis
A flow cytometry-based Hoechst 33342 dye exclusion assay for SP cell analysis was performed as previously described (24). In brief, the shSNTB1 or shVec. transfected cells were harvested, washed twice with cold PBS, and gently resuspended in DMEM medium containing 2% FBS. After staining with the Hoechst 33342 and propidium iodide (PI) to gate viable cells, the cells were filtered through a 40 µm cell strainer before flow cytometric analysis.
TOP/FOP luciferase reporter assay
To assess the transcriptional activity of β-catenin in CRC cells, we employed the TOP/FOP reporter dual luciferase system. Cells were transfected with TOP or FOP flash plasmids and the indicated shRNA-encoding plasmids in a ratio of 3:1. Renilla luciferase was applied as an internal control reporter. Luciferase activity was measured 48 h after transfection and normalized to that of the Renilla luciferase. The luciferase activity of shVec.-treated cells was set as 1 (n=3).
Statistical analysis
All numerical data are presented as mean ± standard deviation (SD) in the three independent experiments. Error bars on all graphs represent the SD between measurements. The relationship between SNTB1 expression and clinicopathological characteristics was analyzed by the chi-square (χ2) test. Survival analysis was carried out using the Kaplan-Meier method together with the log-rank test. One-way analysis of variance (ANOVA) was applied to analyze the statistically significant differences between the means of multiple groups. P<0.05 indicates statistical significance; P<0.01 and P<0.001 are considered as highly significant.
Results
SNTB1 is upregulated in CRC cells and tissues
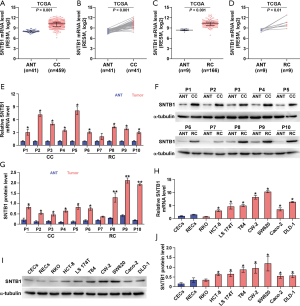
To explore the potential role of SNTB1 in colonic cancer (CC) and RC development, we examined SNTB1 expression using the TCGA RNAseq database. SNTB1 mRNA levels were elevated in cancerous tissues compared to their respective ANT (Figure 1A,B,C,D). We detected SNTB1 abundance in 10 pairs of CRC tumor samples and their respective ANT samples by qRT-PCR and Western blot. Consistent with the above results, the mRNA and protein levels of SNTB1 were upregulated in CRC tissues (Figure 1E,F,G). We also assessed SNTB1 expression in colonic or rectal epithelial cells and eight CRC cell lines. Overall, differential elevations of SNTB1 levels were observed in CRC cell lines compared with normal epithelial cells. Moreover, six of the eight CRC cell lines displayed SNTB1 upregulation in mRNA and protein levels, although the mRNA levels did not correlate well with the corresponding protein levels in individual cell lines (Figure 1H,I,J). These findings indicate that SNTB1 correlates with CRC progression and may be implicated in colorectal tumorigenesis.
SNTB1 expression correlates with CRC clinical prognostic factors
To assess the clinicopathologic significance of SNTB1 in CRC samples, we subsequently used IHC to determine SNTB1 levels in a surgical cohort of CRC patients. Colorectal polyps or cancer issues were characterized as benign (noncancerous), precancerous, or malignant according to the degree of malignancy. As shown in Figure 2A, the benign sample showed no or low SNTB1 expression. In contrast, SNTB1 expression was upregulated in the precancerous sample and significantly increased in more advanced tumor samples (TNM stage I–IV). To further understand the correlation between SNTB1 expression and prognostic factors, we divided the CRC samples into two groups according to the SNTB1 expression level in the tumor tissues: a low SNTB1 expression group (with an immunostaining intensity score ≤4) and a high SNTB1 expression group (with an immunostaining score >4). An immunostaining intensity score of 4 was the median value of all sample tissues. The immunostaining scores of SNTB1 from the benign, precancerous, and malignant samples revealed an overall positive correlation between SNTB1 expression and tumor malignancy and are summarized in Figure 2B.
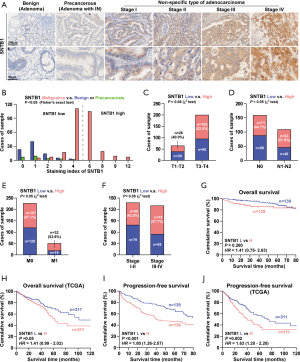
Consistently, we found that SNTB1 expression was closely and positively correlated with advanced tumor stages, including the size of the tumor (T stage, Figure 2C), lymph node status (N stage, Figure 2D), metastasis phenotype (M stage, Figure 2E), and the American Joint Committee on Cancer (AJCC) pathological stages (Figure 2F), implying a correlation between the clinicopathologic characteristics of patients and SNTB1 abundance. We next examined the possible relevance of SNTB1 expression to the overall survival of patients with CRC. While the difference between the high and low SNTB1-expression tissue samples of clinical patients was slight, patients from the TCGA database with a high SNTB1 expression exhibited markedly poorer clinical outcomes than those with a low expression (Figure 2G,H). Notably, patients with a high SNTB1 expression had a lower mean progression-free survival time than the SNTB1-low group (Figure 2I,J). The overall and progression-free survival were inversely related to SNTB1 levels, suggesting a strong correspondence between the levels of SNTB1 expression and the overall survival rate of patients with CRC. Collectively, these studies suggest that STNB1 is a potential indicator of colorectal carcinoma aggressiveness.
SNTB1 silencing suppresses CRC cell proliferation and colony formation in vitro
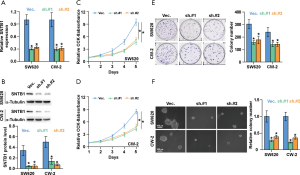
We next sought to evaluate the functional role of SNTB1 on CRC cell proliferation. To this end, two CRC cell lines, SW620 and CW-2, that stably express specific shRNA-SNTB1 were established to knock down the endogenous SNTB1 level by two pairs of small hairpin RNAs. We chose these two cell lines because they expressed the highest levels of the SNTB1 gene in the eight CRC cell lines described above. The mRNA and protein levels of SNTB1 were then determined by qRT-PCR and Western blot. As shown in Figure 3A,B, the SNTB1 expression in the SW620 and CW-2 cells was significantly decreased by approximately 70% compared to the vector control. No noticeable difference in the knockdown efficiency was observed between the two shRNA-SNTB1 variants (sh.#1 and sh.#2). These data suggest that the transcription and translation of the SNTB1 gene were strongly blocked upon shRNA transduction. Applying these established SNTB1-deficient cell lines as models, we performed CCK8 assay and colony-formation experiments to evaluate the effect of SNTB1 on cancer cell growth. The CCK8 assays indicated that SNTB1 silencing robustly impeded the cell proliferation rates in the SW620 and CW-2 cells (Figure 3C,D). Consistently, colony formation assays revealed that SNTB1 silencing repressed colony-generating capability (Figure 3E). The anchorage-independent growth assays demonstrated that SNTB1 knockdown decreased the colony number and size compared to the vector shRNA control group (Figure 3F).
SNTB1 silencing repressed tumor growth in xenograft mice
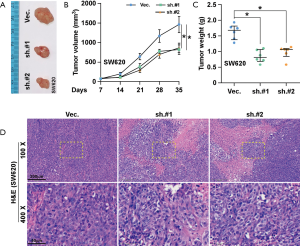
To further investigate the role of SNTB1 on CRC tumorigenesis, SW620 cells transduced with either shVec. or shSNTB1 were injected into nude mice. Phenotypically, xenograft mice with transplanted shSNTB1 tumor cells displayed an alleviated tumor burden compared with mice engrafted with shVec. tumor cells (Figure 4A). SNTB1 silencing robustly suppressed tumor growth at the indicated time (Figure 4B). Moreover, tumors derived from the shSNTB1 group exhibited an approximate 50% reduction of the average tumor weight at sacrifice compared to shVec. mice (Figure 4C). In line with previous data, the result of hematoxylin and eosin (H&E) staining revealed that the proliferative ability of shSNTB1 tumor cells to invade surrounding cells was significantly reduced (Figure 4D). Collectively, these data indicate that suppressing SNTB1 expression inhibits tumorigenesis in vivo.
SNTB1 knockdown attenuated the β-catenin signaling pathway and stemness in CRC
To investigate the molecular mechanism underlying the inhibitory effect of SNTB1 knockdown on CRC cell proliferation, we then applied a gene set enrichment analysis (GSEA) to identify functional gene sets correlated with SNTB1 expression. As indicated in Figure 5A, we found gene sets involved in the β-catenin (CTNNB1) signaling pathway and cell stemness were enriched in the SNTB1-high samples, and the enrichment score (ES) declined in association with reduced SNTB1 expression. The ESs and their corresponding estimation of significance levels (P values) are indicated within the GSEA graphs. This data suggests that SNTB1 expression may be associated with β-catenin activity and cancer cell stemness maintenance. To explore the correlation between the SNTB1 level and β-catenin oncogenic signature, a TOP/FOP flash dual-luciferase reporter assay was performed to evaluate β-catenin activity. As shown in Figure 5B, SNTB1 silencing reduced the transcription of the transfected TOP-Flash reporter by approximately 50% compared with cells transfected with shVec. In contrast, SNTB1 knockdown did not affect the luciferase activity in cells transfected with FOP-Flash plasmids, which contain a mutation in the TCF/LEF binding site. These data indicate that SNTB1 downregulation perturbed the β-catenin-mediated transcriptional activity in CRC cells.
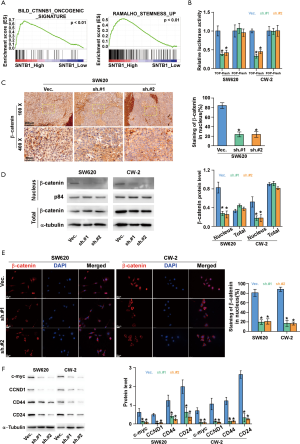
To further validate these observations, we next assessed the association between SNTB1 levels and β-catenin abundance via immunostaining. In the SW620 cell-transplanted xenograft mice, the SNTB1-silenced tumor tissues exhibited a weaker staining intensity of β-catenin in the nucleus (Figure 5C). Western blot analysis also indicated that shSNTB1 transfection robustly blocked the nuclear translocation of β-catenin without affecting its cellular expression level (Figure 5D). Immunofluorescence assay and quantitative analysis confirmed that shSNTB1 knockdown significantly inhibited the amount of β-catenin in the nucleus compared with the control group (Figure 5E). These data suggest that SNTB1 knockdown repressed the nuclear translocation of β-catenin in CRC cells.
The role of the canonical Wnt/β-catenin signaling pathway in cell proliferation has been demonstrated in various types of cancer, including CRC. Aberrant activation of the β-catenin pathway induces accumulation of β-catenin in the nucleus and thereby stimulates the transcription of c-Myc and CCND1, which is a key component within cell cycle checkpoints. We next examined the protein levels of these β-catenin downstream effectors in the context of SNTB1 knockdown. Western blot analysis revealed that SNTB1 silencing robustly reduced the expression of c-Myc and CCND1 in CRC cells compared with the vector shRNA control cells (Figure 5F). These results demonstrated that SNTB1 knockdown represses tumorigenesis through the β-catenin-c-Myc signaling pathway. In addition to modulating the cell proliferation process, the canonical β-catenin pathway is also reported to be critical for cancer stem cell regulation. The cell surface proteins CD24 and CD44 are putative markers for cancer stem cell populations in CRC. Western blot analysis also indicated that SNTB1 knockdown downregulates CD44 and CD24 expressions in SW620 and CW-2 cells (Figure 5F). The reduced protein levels of c-Myc, CCND1, CD44, and CD24 are in line with the GSEA analysis that SNTB1 expression is closely correlated with β-catenin activity and the stemness maintenance of cancer cells.
SNTB1 silencing decreases spherogenicity and inhibits cancer stemness and tumorigenesis
To further examine the functional relevance of SNTB1 in stemness, we assessed the cancer stemness-related gene expressions in SNTB1-silenced CRC cells. As indicated in Figure 6A, the stemness-related genes, including Kruppel like factor 4 (KLF4), EpCAM, aldehyde dehydrogenase 1 member A1 (ALDH1A1), CD133, POU5F1 (POU class 5 homeobox 1, also known as OCT4), and LGR5 were analyzed. shSNTB1 transduction significantly decreased the mRNA expressions of EpCAM, CD133, and LGR5. The results of Western blot analysis confirmed that the translational levels of these three genes were strongly reduced (Figure 6B). The capacity of single CRC cells to form spheres, a hallmark of malignant potential, was investigated in shSNTB1-transduced CRC cells. Silencing of SNTB1 expression significantly impaired sphere formation in two CRC cell lines; the percentage of sphere formation dropped from 24% to around 10% for SW620 cells and from 31% to approximately 14% for CW-2 cells (Figure 6C).
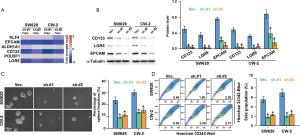
In addition, stem cells are frequently identified as SP cells based on the ABC transporter-mediated efflux of the Hoechst 33342 dye. We evaluated the proportion of the SP within the total cancer cells by flow cytometry-based Hoechst 33342 staining. Figure 6D depicts the Hoechst bright-fluorescence main population (MP) cells and the corresponding low-Hoechst fluorescence SP cells (the tail region of the dot plot) in the SW620 and CW-2 cell lines. The proportion of SP cells with shSNTB1 transduction was decreased by 60–70% compared with those in the shVec. control treatment. These results thus provide evidence that SNTB1 contributes to CRC stemness and β-catenin signaling pathway modulation.
Activation of β-catenin reverses the anti-proliferation effect induced by SNTB1 silencing
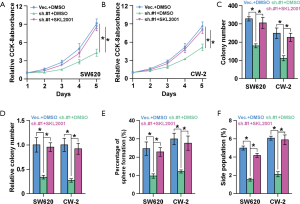
To investigate whether SNTB1 regulates cancer stemness and tumorigenesis through modulation of the β-catenin signaling pathway, we next applied the β-catenin agonist SKL2001 to explore its potential in counteracting SNTB1 silencing. CCK8 assays indicated that SNTB1 knockdown inhibited cell proliferation in CRC cells, but β-catenin agonist SKL2001 treatment rescued cell growth to a level similar to the shVec. controls (Figure 7A,B). The in vitro colony formation assay and anchorage-independent growth assay further suggested that SKL2001 treatment rescued the phenotypes of colony growth inhibition of the SNTB1 knockdown, thereby increasing colony numbers in both SW620 and CW-2 cells (Figure 7C,D). These data suggest that SNTB1 regulates tumor growth through the β-catenin signaling pathway. In addition to cell proliferative capacity, our previous data demonstrated that SNTB1 knockdown impaired tumorigenesis and CRC cell stemness. Sphere formation assays revealed that the β-catenin agonist SKL2001 counteracted the suppressing effect of shSNTB1 on clonogenicity and rescued the percentage of sphere formation to above 20%, a level which is similar to that of “shVec. + DMSO” treatment (Figure 7E). Flow cytometry-based SP cell analysis depicted that shSNTB1 significantly decreased the proportion of SP cells, whereas SKL2001 treatment reversed the effect of shSNTB1 and restored the levels of SP cells in SW620 and CW-2 cells (Figure 7F). By contrast, there was no significant difference in sphere formation or SP percentage between the “shVec. + DMSO” group and the “shSNTB1 + SKL2001” group. Collectively, these results demonstrated that treatment with the β-catenin agonist SKL2001 attenuated the SNTB1-induced inhibitory effect on CRC growth and stemness.
Discussion
The syntrophins are a family of membrane-associated adaptor proteins that bind and regulate dystrophin, serving as scaffold proteins for various transmembrane and intracellular signaling molecules. As one of the least characterized family members, SNTB1 and its physiological role are poorly understood, nevertheless it has biological significance in tumorigenesis.
In this study, we identified aberrant SNTB1 expression in colorectal carcinomas. The elevated level of SNTB1 is positively correlated with malignant clinicopathologic characteristics and the survival of patients with CRC. To further investigate the relevance of SNTB1 expression and tumorigenesis, xenograft mice with SNTB1 knockdown generated by shSNTB1 transduction displayed a tumor volume reduction compared with those treated with vector shRNA. These results suggest that SNTB1 abundance is associated with CRC progression. To the best of our knowledge, no study has explored SNTB1 expression in CRC thus far. A previous study has reported that another syntrophin isoform, SNTA1, is differentially expressed in various human cancers, exhibiting elevated levels in breast cancer but decreased expression in esophageal carcinoma. However, no apparent differences in SNTA1 expression were observed in lung, colon, or rectal cancers (21).
GSEA was performed to explore the molecular mechanism underlying SNTB1’s contribution to CRC progression. Results showed that gene sets regulating the β-catenin pathway and cancer stemness were positively correlated with SNTB1 expression. There is considerable evidence that the Wnt/β-catenin signaling pathway is essential for CRC initiation and progression, and it is one of the leading targets in the development of therapeutic interventions for CRC. Moreover, the β-catenin pathway is frequently upregulated in CRC and is reported to be critical for the maintenance of cancer stem cell properties (28,29). Herein, a shRNA-based SNTB1 silencing approach was established to investigate the role of SNTB1 in CRC cells. We showed that SNTB1 knockdown significantly repressed the nuclear translocation of β-catenin without affecting its cellular expression level. The downregulation of β-catenin in nuclear would impede the transcription of its downstream effectors like c-Myc and CCND1, negatively regulate their expression in CRC. c-Myc and CCND1 are two core components in modulating cell cycle and cellular proliferation. Our data suggested that SNTB1 promotes CRC tumorigenesis and progression by regulating the β-catenin signaling pathway. And importantly, SNTB1 knockdown suppressed the expression of cancer stem cell surface markers and reduced the percentage and clonogenicity of SP cells. These findings were endorsed by rescue experiments using the β-catenin agonist SKL2001, which counteracts the phenotypes induced by SNTB1 silencing. In particular, SKL2001 reversed the suppressive effect of SNTB1 silencing on tumor growth and restored the level of SP cells in CRC. These findings provide insights for a better understanding of the physiological role of SNTB1 and how SNTB1 intertwines with the β-catenin signaling pathway to regulate tumor growth.
In summary, our findings reveal a previously unappreciated role of SNTB1 in the pathogenesis and progression of CRC. The association of SNTB1 expression and CRC malignancy identified in this study points to the potential value of SNTB1 as a molecular marker for CRC prognosis. Our study also provides an insight into the mechanistic link between SNTB1 and β-catenin signaling. SNTB1 is found, at least partially, to prompt tumor growth and retain stemness through β-catenin signaling activation. Nevertheless, the exact mechanism of SNTB1 in β-catenin activation and the consequent stimulation of CRC progression has not yet been fully explored. Future studies will be needed to discover any missing pieces of the puzzle and elucidate how SNTB1 mediates tumor progression in CRC.
Acknowledgments
Funding: This study was supported by grants from the Dongguan Social Science and Technology Development Project (201950715025192); the Guangdong Basic and Applied Basic Research Foundation (2019A1515110042, 2019A1515011713, 2019A1515110357, 2021A1515012054), Discipline Construction Project of Guangdong Medical University, Shenzhen Basic Research Program (JCYJ20180306173853283), the Research Foundation of Guangdong Medical University for Ph.D. Staff (GDMUB2019038, GDMUB2020017), the Key Cultivation Project of Guangdong Medical University (GDMUZ2019001), the Science and Technology Program of Zhanjiang (2020A01028), the Administration of Traditional Chinese Medicine of Guangdong Province (20211217), and the Medical Science Foundation of Guangdong Province (A2020211).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2700
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2700
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2700). Dr. JZ reports funding from Guangdong Basic and Applied Basic Research Foundation, Dongguan Social Science and Technology Development Project, Shenzhen Basic Research Program, and the Key Cultivation Project of Guangdong Medical University. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval and informed consent for the collection of specimens was endorsed by the Institutional Research Ethics Committee of the Affiliated Jiangmen Hospital of Sun Yat-sen University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Animal experiments in the study were approved by the Animal Experimental Ethics Committee of Guangdong Medical University (GDY1902070) in compliance with the National Guidelines for the Care and Use of Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8:511-24. [Crossref] [PubMed]
- O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106-10. [Crossref] [PubMed]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 2007;104:10158-63. [Crossref] [PubMed]
- Kobayashi S, Yamada-Okabe H, Suzuki M, et al. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells 2012;30:2631-44. [Crossref] [PubMed]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004;20:781-810. [Crossref] [PubMed]
- Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol 2003;129:199-221. [Crossref] [PubMed]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005;434:843-50. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget 2017;8:33972-89. [Crossref] [PubMed]
- van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002;111:241-50. [Crossref] [PubMed]
- Ou CY, Kim JH, Yang CK, et al. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and beta-catenin and for anchorage-independent growth of human colon carcinoma cells. J Biol Chem 2009;284:20629-37. [Crossref] [PubMed]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999;398:422-6. [Crossref] [PubMed]
- He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281:1509-12. [Crossref] [PubMed]
- Bryja V, Červenka I, Čajánek L. The connections of Wnt pathway components with cell cycle and centrosome: side effects or a hidden logic? Crit Rev Biochem Mol Biol 2017;52:614-37. [Crossref] [PubMed]
- de Sousa EM, Vermeulen L, Richel D, et al. Targeting Wnt signaling in colon cancer stem cells. Clin Cancer Res 2011;17:647-53. [Crossref] [PubMed]
- Lukaszewicz AI, Nguyen C, Melendez E, et al. The mode of stem cell division is dependent on the differential interaction of β-catenin with the Kat3 coactivators CBP or p300. Cancers (Basel) 2019;11:962. [Crossref] [PubMed]
- Pandit H, Li Y, Li X, et al. Enrichment of cancer stem cells via β-catenin contributing to the tumorigenesis of hepatocellular carcinoma. BMC Cancer 2018;18:783. [Crossref] [PubMed]
- Yamashita T, Budhu A, Forgues M, et al. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res 2007;67:10831-9. [Crossref] [PubMed]
- Bhat SS, Ali R, Khanday FA. Syntrophins entangled in cytoskeletal meshwork: helping to hold it all together. Cell Prolif 2019;52:e12562 [Crossref] [PubMed]
- Neely JD, Amiry-Moghaddam M, Ottersen OP, et al. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc Natl Acad Sci U S A 2001;98:14108-13. [Crossref] [PubMed]
- Bhat HF, Baba RA, Bashir M, et al. Alpha-1-syntrophin protein is differentially expressed in human cancers. Biomarkers 2011;16:31-6. [Crossref] [PubMed]
- Bhat HF, Adams ME, Khanday FA. Syntrophin proteins as Santa Claus: role(s) in cell signal transduction. Cell Mol Life Sci 2013;70:2533-54. [Crossref] [PubMed]
- Compton AG, Cooper ST, Hill PM, et al. The syntrophin-dystrobrevin subcomplex in human neuromuscular disorders. J Neuropathol Exp Neurol 2005;64:350-61. [Crossref] [PubMed]
- Ren D, Lin B, Zhang X, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017;8:49807-23. [Crossref] [PubMed]
- Zhang X, Zhang L, Lin B, et al. Phospholipid Phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable Cationic Channel in lung carcinoma cells. Mol Cancer 2017;16:147. [Crossref] [PubMed]
- Ye Z, Liang Y, Ma Y, et al. Targeted photodynamic therapy of cancer using a novel gallium (III) tris (ethoxycarbonyl) corrole conjugated-mAb directed against cancer/testis antigens 83. Cancer Med 2018;7:3057-65. [Crossref] [PubMed]
- Wang B, Liang Y, Chai X, et al. Ectodysplasin A receptor (EDAR) promotes colorectal cancer cell proliferation via regulation of the Wnt/β-catenin signaling pathway. Exp Cell Res 2020;395:112170 [Crossref] [PubMed]
- Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006;25:7531-7. [Crossref] [PubMed]
- Bahrami A, Amerizadeh F. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: rational and progress. J Cell Biochem 2017;118:1979-83. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


