Abstract
The aim of this study was to evaluate the clinicopathological and prognostic relevance of PIK3CA mutations in Chinese patients with surgically resected cervical cancer. PIK3CA mutations were screened in 771 cervical cancer specimens using reverse transcription polymerase chain reaction and Sanger sequencing. In total, 13.6% (105 of 771) of patients harbored non-synonymous PIK3CA mutations. Patients harboring PIK3CA mutations were older than patients with wild-type PIK3CA (mean age: 50.7 years vs. 47.0 years, P < 0.01). PIK3CA mutations were more commonly observed in postmenopausal patients than in premenopausal patients (19.6% vs. 10.2%, P < 0.01). PIK3CA mutations were more common in squamous cell carcinomas than in non-squamous cell tumors (15.3% vs 7.3%, of P < 0.01). The 3-year relapse-free survival was 90.2% for PIK3CA mutant patients and 80.9% for PIK3CA wild-type patients (P = 0.03). PIK3CA mutation was confirmed as an independent predictor for better treatment outcome in the multivariate analyses (HR = 0.54, 95% CI: 0.29–0.99, P = 0.048). PIK3CA mutations were significantly associated with less distant metastases (mutant-type: 8/105, wild-type: 98/666, p = 0.048). Thus, patients with mutant PIK3CA had distinct characteristics in age, menopausal status and histological subtype and have better treatment outcome and less distant metastasis after surgery-based multimodal therapy.
Similar content being viewed by others
Introduction
Cervical cancer is the eighth deadliest cancer and is responsible for over 20,000 deaths in China annually1. Despite significant improvements in screening, diagnosis and therapy of cervical cancer over the past decade, progress in the management of advanced/recurrent cervical cancer has been underwhelming, as the median overall survival in advanced/recurrent cervical cancer patients remains between 10 and 13 months2.
Genes involved in the PI3K pathway represent potential therapeutic targets for cancers and PIK3CA mutation status may be useful as a biomarker for targeted therapy of cervical cancer. The frequently mutated and constitutively activated PI3K pathway is involved in the pathogenesis of various human cancers. The PI3K pathway is an attractive cancer therapeutic target. PI3K pathway inhibitors, such as buparlisib, everolimus and temsirolimus, have demonstrated clinical efficacy in several different cancers3,4,5,6. PIK3CA is one of the most commonly mutated genes associated with cervical cancer7. Most PIK3CA mutations found in cervical cancer occur in the helical domain (exon 9), whereas a few PIK3CA mutations have been reported to affect the kinase domain (exon 20)8. Preclinical and clinical studies have demonstrated that PIK3CA mutations may predict tumor response to PI3K pathway inhibitors9,10,11,12. Therefore, the identification and characterization of cervical cancer patients harboring mutant PIK3CA are important for designing clinical trials investigating PI3K pathway inhibitors.
However, the clinicopathological characteristics and prognostic impact of PIK3CA-mutated cervical cancer have not been well established. It is necessary to conduct PIK3CA mutational analyses in a large cohort of cervical cancer patients. Therefore, in this study, we investigated the clinicopathological and prognostic relevance of PIK3CA mutations in a large cohort of Chinese patients with surgically resected cervical cancer.
Results
PIK3CA mutation in cervical cancers
The prevalence of PIK3CA mutations in our cohort of 771 patients with FIGO stage IB/IIA cervical cancer was determined by RT-PCR–based direct sequencing. Nonsynonymous PIK3CA mutations were found in 105 cases (13.6%) and all of these mutations were base substitutions. Ten mutations occurred in the helical domain (HD), whereas 95 mutations occurred in the kinase domain (KD). No tumors harbored mutations in both the HD and KD. In total, 94.3% (99) of the 105 mutations occurred at previously identified hotspots, including 62 amino acid substitutions at residue 545, 32 amino acid substitutions at residue 542 and 5 amino acid substitutions at residue 1,047. The most common mutations were E545K and E542K, both of which affect exon 9, which were found in 59 and 32 samples, respectively. H1047R mutation, which has been reported to predict an increased response to PI3K/AKT/mTOR signaling pathway inhibitors, was found in 4 cases13. Several rare nonsynonymous base substitutions were identified in our study, some of which have been reported in the Catalog of Somatic Mutations in Cancer (COSMIC) database. Two mutations identified in our study were not previously reported. One hundred mutations (95.2%) were recognized as activating mutations, whereas the effects of the remaining five mutations are unknown (Table 1).
Clinicopathological association of PIK3CA mutations
The 771 patients in our study comprised 606 (78.6%) patients with squamous cell carcinomas (SCCs), 101 (13.1%) patients with adenocarcinomas (ACs), 44 (5.7%) patients with adenosquamous carcinomas (ASCs) and 20 (2.6%) patients with other rare histopathological subtypes. Two-hundred two (26.2%) patients presented with pelvic or paraaortic lymph node metastasis during pathologic examination of the surgical specimens. Five-hundred forty (70.0%) patients underwent post-operative radiotherapy or chemoradiotherapy for a high-risk for recurrence.
Patient age, menopausal status and histological subtypes were strong predictors of PIK3CA mutation status. Patients harboring PIK3CA mutations were older than patients with wild-type PIK3CA (mean 50.7 vs. 47.0 years; P < 0.01) (Table 2). PIK3CA mutations were identified in 23.5% of patients aged ≥60 years, 17.5% of patients aged 50 to 59 years, 10.9% of patients aged 40 to 49 years and 8.5% of patients aged <40 years. This trend was significantly different when compared with PIK3CA wild-type cohort (P < 0.01). PIK3CA mutations were more commonly observed in postmenopausal patients than in premenopausal patients (19.6% vs. 10.2%, P < 0.01). In addition, PIK3CA mutations occurred more frequently in squamous cell carcinomas than in non-squamous cell tumors (15.3% vs. 7.3%, P = 0.01). In this study, PIK3CA mutation did not correlate with other clinicopathonogical features, such as clinical FIGO stage, node metastasis, tumor size, myometrial invasion, LVSI and parametrial involvement (Table 2).
Prognostic role of PIK3CA mutation
In the median follow-up of 38 months (range: 1–59 months), 142 patients experienced distant metastases (n = 106) or pelvic recurrence (n = 36). PIK3CA mutations were significantly associated with distant metastases (mutant-type: 8/105, wild-type: 98/666, p = 0.048), whereas, the correlation between PIK3CA mutation status and pelvic recurrence was not confirmed in this cohort of patients (mutant-type: 3/105, wild-type: 33/666, p = 0.459).
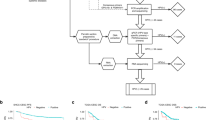
Univariate analysis revealed a striking association between PIK3CA mutations and patient survival. Patients harboring mutant PIK3CA showed a better treatment outcome than patients with wild-type PIK3CA. The 3-year relapse-free survival (RFS) rate was 90.2% for PIK3CA mutant patients and 80.9% for PIK3CA wild-type patients (P = 0.03) (Fig. 1A and Table 3). Our multivariate analyses revealed that PIK3CA mutation in cervical cancer was an independent predictor for better RFS (HR = 0.54, 95% CI: 0.29–0.99, P = 0.048) (Table 3).
To exclude confounding effects associated with histological subtypes, a survival analysis was conducted in patients with squamous cell carcinomas. A similar dichotomized RFS trend was observed according to PIK3CA mutation status in patients with squamous cell carcinomas; however, the difference did not reach statistical significance (3-RFS: 91.3% for mutant PIK3CA vs. 83.5% for wild-type PIK3CA, P = 0.07), which could be due to the short follow-up duration and the limited number of patients experiencing disease relapse (Fig. 1B). We did not observe enough recurrent cases (2 relapses in 12 cases with PIK3CA mutations) to effectively evaluate the association between PIK3CA mutation status and RFS in the subgroups of patients with non-squamous cell tumors (shown in supplementary Figure S1A).
A survival association analysis was performed in 540 patients who underwent postoperative radiotherapy. In this group of patients, better treatment outcomes were observed in patients harboring mutant PIK3CA than in patients with wild-type PIK3CA (3-year RFS: 89.1% vs. 76.0% P = 0.02). In patients who did not receive adjuvant therapy, no significant difference was observed in the 3-year RFS based on the presence or absence of PIK3CA mutation (shown in supplementary Figure S1B).
In order to explore which specific mutations in PIK3CA would be really useful for prognosis, all the PIK3CA mutants were subdivided into three groups according to different mutated position of amino acid residuals (pos.542, pos.545 and other positions). Similar trends of RFS were observed in the three subgroups of patients, while only the subgroup of patients with mutations on pos.542 was confirmed to have better RFS than the patients with wide-type PIK3CA (p = 0.025). The differences of RFS between the patients with mutations on the other positions and those with wide-type did not reach statistical significance probably due to limited cases (p = 0.366 for pos. 545 and p = 0.379 for other positions, supplementary Figure S2A). Furthermore, PIK3CA mutations were subdivided according to different exons (exon 9 and exon 20). Similar trends of RFS were observed in the two subgroups. However, only the patients with mutations on exon 9 had better survival than the patients with wide-type (p = 0.040). The difference between the subgroups with mutations on exon 20 and without mutations did not reached statistical significance probably due to limited cases number (p = 0.443, shown in supplementary Figure S2B).
Discussion
The mutation status in the PIK3CA gene could be used as a targeted biomarker for cervical cancer patients. Clinicopathological and prognostic features of PIK3CA mutations in tumors remain inconclusive. Although several previous studies have attempted to investigate this association14,15,16,17,18, most of these studies are underpowered for robust statistical analyses because of small sample size (less than 200 cases), selection bias or a lack of control for confounding factors. Our study is the first large cohort study to investigate the clinicopathological and prognostic relevance of PIK3CA mutations in patients with surgically resected cervical cancer.
The prevalence (13.6%) of PIK3CA mutations in our study is consistent with that reported in the COSMIC database (10%) as well as with a whole-exome sequencing study (14%)7. However, our reported prevalence is also substantially lower than in other investigations (23–33%)10,14,15. The reasons for the differences between these studies and ours may include ethnic heterogeneity, inclusion of late-stage cases and a greater probability of contamination during specimen processing for paraffin-embedded tissues than for fresh tissues. Importantly, our study is consistent with previous investigations in that most of the PIK3CA mutations clustered within exon 9. This distribution is distinct in cervical cancers when compared with other types of tumors that present with frequent PIK3CA mutations, such as breast cancer19 and endometrial cancer20. Different mutation hotspots in PIK3CA may predict different responses to inhibitors during therapy13.
PIK3CA mutations occurred more frequently in older and postmenopausal patients in our study, which is in agreement with a study performed by Cui et al.17. In addition, the histological subtype can serve as another predictor for PIK3CA mutation in cervical cancer. Similar to our results, Alexi et al. also observed a higher mutation rate in SCC samples than in AC samples, although, in their study, the difference did not reach statistical significance, which is most likely due to the limited number of cases included in their analyses14. No significant differences in the mutation frequencies were observed in tumor subgroups according to other clinical characteristics.
The association between PIK3CA mutation status and prognosis was inconclusive. In a study performed by McIntyre et al., PIK3CA mutation is associated with a worse overall survival in patients with FIGO stage IB/II cervical cancer treated with radical chemoradiotherapy15. In contrast, a study performed by Hou et al. suggested that PIK3CA mutation is associated with a significantly longer overall survival in patients with metastatic or recurrent cervical squamous cell carcinoma10. Wright et al. reported that PIK3CA mutations were associated with shorter survival in unspecified cervical cancers14. In our study of a large cohort of Chinese patients who underwent surgery-based multimodal therapy, PIK3CA mutations were associated with a longer RFS on both univariate analysis and multivariate analysis including almost all risk factors for recurrence (pathological subgroup, FIGO stage, lymph node metastasis, parametrial involvement, lymph vascular involvement and deep myometrial invasion).
There were some studies which addressed that PIK3CA mutation associated with treatment outcomes in cervical cancer10,14,15,21, however, the sample sizes were too small to draw a conclusion. In this study of a large cohort of Chinese patients who underwent surgery-based multimodal therapy, PIK3CA mutations were associated with a longer RFS on both univariate analysis and multivariate analysis including various risk factors for recurrence. On the contrary, PIK3CA mutation was reported to be associated with poor response or poor survival after definitive radiotherapy by McIntyre et al.15 and De la Rochefordiere et al.21. Furthermore, it had been observed that constitutively activated PI3K pathway promoted resistance to radiation and inhibitors of PI3K pathway radiosensitized human cervical cancer cell lines22,23. The reasons for the differences between our study and their studies were as follows: (1) all the tumors in this study were surgically resected cancers staged as FIGO IB1-IIA2, relatively earlier stage than the tumors treated with primary definitive radiotherapy in the other studies; (2) In this study, all the patients received primary radical surgeries and a considerable proportion of patients received adjuvant radiation after radical surgery. After radical surgery, there were no visible residual tumors in patient’s body, which would minimize the impact of PIK3CA mutation on radioresistance; (3) We observed higher risk of distant metastases after treatment in PIK3CA wild-type cohort (14.7%) than PIK3CA mutant cohort (7.6%) (p = 0.048). Although the reason for this is not clear, similar results have been reported in breast cancer. Sabine et al. reported that patients with breast cancer without PIK3CA mutations were at a significantly higher risk of distant metastases than patients whose tumors harbored PIK3CA mutation24.
This study contains certain limitations. First, given that HPV genotyping is not routinely analyzed for cervical cancer patient management, HPV genotyping analysis was not included in this study. Second, whether the rare mutant variants observed in our study could activate the PI3K pathway requires functional verification. Third, due to the relatively short follow-up period and the small proportion of the patients dying from cervical cancer, the overall survival (OS) was not calculated in this study. Furthermore, because of the limited number of cases with a mutation in exon 20, we were unable to compare clinical features between patients with PIK3CA mutations in exon 9 and patients with PIK3CA mutations in exon 20.
In summary, our data demonstrate that a considerable proportion of Chinese cervical cancer patients harbor mutant PIK3CA. Patients with mutant PIK3CA tumors show significant differences when compared with patients with PIK3CA wild-type tumors with respect to age, menopausal status, histological subtype, treatment outcome and recurrent pattern after surgery-based multimodal therapy.
Methods
Patients and specimens
This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (FUSCC 050432-4-1212B) and was carried out in accordance with the approved guidelines. All patients provided written informed consent. In total, 771 Chinese women were included in this study. Each of them satisfied the following criteria: pathologically confirmed primary cervical carcinoma, FIGO stage IB1-IIA2 disease, no neoadjuvant chemotherapy or radiotherapy, having undergone radical surgery for the primary tumor and no pre-operative conization. The tumor specimens were prepared and processed as previously described8. Cervical tumor specimens were collected during radical hysterectomy or trachelectomy procedures between January 2010 and December 2012. Histological assessment of the specimens was performed by two independent pathologists (Xuxia Shen and Wentao Yang) and only specimens with more than 50% tumor cells were included in our analysis. The tumor specimens were preserved in RNAlater solution (Ambion) and stored at −80 °C until analyzed. Clinicopathological data were prospectively retrieved from patient medical records. The patients were followed up regularly until relapse occurred.
Mutation analysis
PIK3CA mutation analysis was conducted without prior knowledge of patient clinicopathological and follow-up data. Total RNA extracted from the tumor tissues was reverse-transcribed into single-stranded cDNA using an M-MLV reverse transcriptase kit (Invitrogen). The helical domain (HD) and the kinase domain (KD) of PIK3CA was PCR amplified with KOD-Plus-Neo DNA polymerase (Toyobo) using the following primers: HD-F: TTCAGCAGTGTGGTAAAGTTCC; HD-R: TACCAAGCAATACATCTGGGCTAC; KD-F: CTGGATACTGTGTAGCTACCTT; KD-R: ATGGATTGTGCAATTCCTATG. The PCR products were directly sequenced from both ends using Sanger sequencing. All mutations were confirmed by an additional independent PCR.
Statistical analysis
The relevance between PIK3CA mutations and clinicopathological characteristics was analyzed by Student’s t-test, Pearson’s chi-square test or Fisher’s exact test. Relapse-free survival (RFS) was defined as the period of time from surgery to the first local, lymph node or distant recurrence. Distant metastasis was defined as any relapse outside of pelvic cavity. Patients who died without evidence of disease were censored. Due to the short duration of patient follow-up, the overall survival (OS) was not calculated in this study. RFS was assessed using the Kaplan–Meier method and differences between the groups were analyzed using the log-rank test. Hazard ratios (HRs) were calculated using Cox proportional-hazards models. The two-sided significance level was set at P < 0.05. Data were analyzed using IBM SPSS Statistics 19 (IBM).
Additional Information
How to cite this article: Xiang, L. et al. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci. Rep. 5, 14035; doi: 10.1038/srep14035 (2015).
Change history
05 October 2015
The version of this Article previously published incorrectly indicated that Gong Yang, Xiaohua Wu and Huijuan Yang contributed equally to the work, rather than Libing Xiang and Wei Jiang. These errors have now been corrected in both the PDF and HTML version of the paper. In addition, Gong Yang, Xiaohua Wu and Huijuan Yang were not listed as corresponding authors in the HTML version of this paper. Correspondence and request for materials should be addressed to yanggong@fudan.edu.cn, docwuxh@hotmail.com and huijuanyang@hotmail.com. This has now been corrected in the HTML.
References
Chen, W. et al. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 26, 48–58 (2014).
Lorusso, D., Petrelli, F., Coinu, A., Raspagliesi, F. & Barni, S. A systematic review comparing cisplatin and carboplatin plus paclitaxel-based chemotherapy for recurrent or metastatic cervical cancer. Gynecol Oncol. 133, 117–123 (2014).
Andre, F. et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 15, 580–591 (2014).
Hess, G. et al. Phase III study to evaluate temsirolimus compared with investigator’s choice therapy for the treatment of relapsed or refractory mantle cell lymphoma. J Clin Oncol. 27, 3822–3829 (2009).
Motzer, R. J. et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 372, 449–456 (2008).
Mayer, I. A. et al. Stand up to cancer phase Ib study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in estrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 32, 1202–1209 (2014).
Ojesina, A. I. et al. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014).
Xiang, L. B. et al. Comprehensive Analysis of Targetable Oncogenic Mutations in Chinese Cervical Cancers. oncotarget (2015, in press).
Janku, F. et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 30, 777–782 (2012).
Hou, M. M. et al. Targeted PI3K/AKT/mTOR therapy for metastatic carcinomas of the cervix: A phase I clinical experience. Oncotarget 5, 11168–11179 (2014).
Spoerke, J. M. et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res. 18, 6771–6683 (2012).
Lopez, S. et al. Taselisib, a selective inhibitor of PIK3CA, is highly effective on PIK3CA-mutated and HER2/neu amplified uterine serous carcinoma in vitro and in vivo. Gynecol Oncol. 135, 312–317 (2014).
Janku, F. et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 73, 276–284 (2013).
Wright, A. A. et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 119, 3776–3783 (2013).
McIntyre, J. B. et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. 128, 409–414 (2013).
Janku, F. et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One 6, e22769 (2011).
Cui, B. et al. Mutation of PIK3CA: possible risk factor for cervical carcinogenesis in older women. Int J Oncol. 34, 409–416 (2009).
Miyake, T. et al. PIK3CA gene mutations and amplifications in uterine cancers, identified by methods that avoid confounding by PIK3CA pseudogene sequences. Cancer Lett. 261, 120–126 (2008).
Sabine, V. S. et al. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J Clin Oncol 32, 2951–2958 (2014).
Rudd, M. L. et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 17, 1331–1340 (2011).
de la Rochefordiere, A. et al. PIK3CA Pathway Mutations Predictive of Poor Response Following Standard Radiochemotherapy +/− Cetuximab in Cervical Cancer Patients. Clin Cancer Res 21, 2530–2537 (2015).
Brognard, J. et al. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res 61, 3986–3997 (2001).
Lee, C. M. et al. Phosphatidylinositol 3-kinase inhibition by LY294002 radiosensitizes human cervical cancer cell lines. Clin Cancer Res 12, 250–25 (2006).
Sabine, V. S. et al. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J Clin Oncol 32, 2951–2958 (2014).
Acknowledgements
This study was supported by a grant from the National Natural Science Foundation of China (30973426)
Author information
Authors and Affiliations
Contributions
Conception and design: X.L., J.W., Y.G., W.X. and Y.H. Acquisition of data: X.L., J.W., L.J., S.X., Y.W., Y.G., W.X. and Y.H. Analysisi and interpretation of data: X.L., J.W., L.J., S.X., Y.W., Y.G., W.X. and Y.H. Manuscript preparation: X.L., J.W., Y.G., W.X. and Y.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xiang, L., Jiang, W., Li, J. et al. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci Rep 5, 14035 (2015). https://doi.org/10.1038/srep14035
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep14035
This article is cited by
-
Integrated genomic and transcriptomic analysis reveals the activation of PI3K signaling pathway in HPV-independent cervical cancers
British Journal of Cancer (2024)
-
First-in-human phase Ia study of the PI3Kα inhibitor CYH33 in patients with solid tumors
Nature Communications (2022)
-
Mutational profiles of marker genes of cervical carcinoma in Bangladeshi patients
BMC Cancer (2021)
-
TP53 mutants and non-HPV16/18 genotypes are poor prognostic factors for concurrent chemoradiotherapy in locally advanced cervical cancer
Scientific Reports (2021)
-
Clinical implication of oncogenic somatic mutations in early-stage cervical cancer with radical hysterectomy
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.