Abstract
Telomerase activity in human cells closely correlates with the expression of its catalytic subunit, telomerase reverse transcriptase (hTERT). Previously, we reported the lack of telomerase activity in normal human oral fibroblasts (NHOF) and the diminution of telomerase activity during senescence in normal human oral keratinocytes (NHOK). To investigate the underlying mechanisms of telomerase regulation in both cell types, we analysed the expression, promoter activity, and methylation status of the hTERT gene. The expression of hTERT mRNA diminished in senescent NHOK, but was not detected in NHOF at any stage of replication. An exogenous hTERT promoter was active in NHOF and in senescing NHOK, indicating that the lack of hTERT gene expression resulted from alteration of the endogenous hTERT promoter. Since methylation is involved in the silencing of numerous genes, we carried out DNA methylation assays. The assay revealed that the hTERT promoter was hypermethylated in NHOF and was gradually methylated during senescence in NHOK. Treatment of NHOF and senescent NHOK with the demethylating agent 5-aza-2′-deoxycytidine restored the expression of endogenous hTERT mRNA. Our results suggest that hypermethylation of the hTERT promoter plays a critical role in the negative regulation of telomerase activity in normal human oral cells.
Similar content being viewed by others
Main
Telomerase is complex, composed of a ribonucleoprotein, reverse transcriptase catalytic subunit, hTERT, template, hTR, and several associated protein cofactors (Feng et al, 1995; Harrington et al, 1997; Meyerson et al, 1997; Nakamura et al, 1997). While hTERT is not expressed in most normal human somatic tissues, hTR is expressed constitutively in most human tissues (Kim et al, 1994; Shay and Bacchetti, 1997). In normal human cells and in tumour cells, telomerase activity is positively correlated with the expression of hTERT (Kim et al, 1994; Shay and Bacchetti, 1997). Therefore, regulation of hTERT expression appears to control telomerase activity. The hTERT promoter region has been cloned and characterised (Horikawa et al, 1999; Takakura et al, 1999), permitting analysis of the molecular mechanisms involved in its regulation (Cong et al, 1999; Horikawa et al, 1999; Takakura et al, 1999; Wick et al, 1999). Transcription factors c-myc, Sp1, AP2, AP4, and c-myb act as activators and several repressors, such as WT1, p53, MZF-2, and Mad1, have been identified (reviewed by Poole et al, 2001). Analysis of the hTERT promoter region also revealed the presence of CpG islands containing CG-rich methylation sites (Cong et al, 1999; Horikawa et al, 1999; Takakura et al, 1999; Wick et al, 1999). Promoter region hypermethylation associated with transcriptional loss is an alternative mode of gene silencing for several tumour-suppressor genes (Zingg and Jones, 1997; Baylin et al, 1998; Herman, 1999). The hypermethylation status of the hTERT promoter has been examined in a variety of human normal and cancer cells (Devereux et al, 1999; Dessain et al, 2000; Guilleret et al, 2002). In these studies, hTERT promoter methylation was observed in telomerase-negative, as well as in telomerase-positive cells, suggesting different modes of regulation of hTERT expression in methylation-dependent and -independent pathways.
Previously, we reported that replicating normal human oral keratinocytes (NHOK) expressed telomerase activity, which gradually disappeared during senescence, and that normal human oral fibroblasts (NHOF) lacked telomerase activity at all stages of replication (Kang et al, 1998). To investigate the mechanism of telomerase inactivation in NHOK and NHOF, we analysed the changes in hTERT expression, hTERT promoter activity, and methylation status of the hTERT promoter, in serially subcultured NHOK and in replicating NHOF. In our present study, we found that the inactivity of telomerase in NHOF and during NHOK senescence was not caused by absence or defective necessary cofactors, but by hypermethylation of CpG islands in the hTERT gene promoter.
Materials and methods
Cells and culture conditions
Primary cultures of NHOF were established from explants of the gingival connective tissue excised from patients undergoing oral surgery. The cells that proliferated outwardly from the explant culture were continuously cultured in 100-mm culture dishes, in DMEM/medium 199 (4 : 1) containing foetal bovine serum (Gemini Bioproducts, Calabasas, CA, USA), and gentamicin (50 μg ml−1). Primary NHOK were prepared from separated epithelial tissue, and serially subcultured in keratinocyte growth medium (KGM, Clonetics, Charlotte, NC, USA) containing 0.15 mM Ca2+, as described previously (Kang et al, 1998). Adenovirus transformed human embryonic kidney cells 293, obtained from the American Type Culture Collection (Manassas, VA, USA), were cultured in DMEM/medium 199 (4 : 1) containing foetal bovine serum (Gemini Bioproducts) and gentamicin (50 μg ml−1). SCC-4 (human squamous cell carcinoma cell line) was obtained from American Type Culture Collection (Manassas, VA, USA), and cultured in DMEM/Ham's F-12 containing 10% foetal bovine serum (Gemini Bioproducts) and gentamicin (50 μg ml−1).
Cell population doublings (PD) were determined after subculturing to postmitotic stage. At the end of each passage, PD was calculated by the formulation, 2N=(Cf/Ci), where N denotes PD, Cf the total cell number harvested at the end of a passage, and Ci the total cell number of attached cells at seeding. The PD time was calculated by dividing the duration of culture in hours by the number of PD.
Analysis of telomerase activity
Cellular extracts were prepared by using CHAPS (lysis buffer) provided from the TRAP-eze Telomerase Detection Kit (Intergen Norcross, GA, USA), as recommended by the manufacturer. Telomerase activity was determined using the TRAP-eze Telomerase Detection Kit as described previously (Kang et al, 1998). Each TRAP reaction contained a cellular extract equivalent to 1 μg protein. The PCR products were electrophoresed in 12.5% nondenaturing polyacrylamide gels, and the radioactive signals were detected by PhosphorImage (Molecular Dynamix, Sunnyvale, CA, USA).
Analysis of endogenous hTERT expression
Total RNA was isolated from the cells using Trizol™ reagent (Invitrogen Carlsbad, CA, USA) and further purified through RNeasy columns (Qiagen, Chatsworth, CA, USA). Following isolated RNA solution in 7.5 μl H2O, the reverse transcription (RT) reaction was performed in first-strand buffer (Invitrogen) containing 200 U Superscript II (Invitrogen), 40 U RNase inhibitor (Perkin-Elmer, Foster City, CA, USA), 10 μ M dithiothreitol, 250 ng random hexamer (Perkin-Elmer), and 2.5 μ M dNTP. The annealing reaction was carried out for 10 min at 25°C, and cDNA synthesis was performed for 50 min at 42°C, followed by 10 min incubation at 70°C for enzyme inactivation.
To amplify hTERT cDNA, PCR reaction was performed with 1 μl RT product using the primers 5′-GCCTGAGCTGTACTTTGTCAA-3′ (forward) and 5′-CGCAAACAGCTTGTTCTCCATGTC-3′ (reverse). The primer set amplified hTERT mRNA species encoding functional full-length (457 bp) or defective spliced isoforms (421, 275, or 239 bp) (Ding et al, 2002). PCR products were separated in 2% agarose gels. The hTERT signals were further amplified by Southern hybridisation using 32P-[dCTP]-radiolabelled probe synthesised from hTERT cDNA, as described in detail previously (Kang et al, 2002).
Analysis of hTERT promoter activity
A pGL3B-TRTP containing a 1670 bp fragment (−1665 to 5) of full-length hTERT promoter upstream of the firefly luciferase gene in the pGL3-basic (Promega Madiso, WI, USA), kindly provided by Dr J Carl Barrett (National Institute of Environmental Health Science), was used for the hTERT promoter assay. Prior to transfection, a six-well plate with approximately 5 × 104 cells per well was inoculated and cultured for 24 h. The pGL3-basic (2 μg well−1) or the pGL3B-TRTP vectors (2 μg well−1) were transfected using Lipofectin reagent (BRL Life Technologies, Inc., Gaithersberg, MD, USA). For better comparison among cells with different transfection efficiencies, the pGL3-control plasmid (2 μg well−1; Promega), which has the firefly luciferase gene under the transcriptional control of SV40 enhancer/promoter, was also transfected into each cell and used for normalisation of the activities shown by the pGL3B-TRTP construct. Cells were collected 48 h after transfection and cell lysates prepared according to Promega's instruction manual. Luciferase activity was measured using a luminometer (Promega).
Methylation analysis of hTERT promoter
Methylation of the hTERT promoter was studied using methylation-specific PCR with bisulphite-modified DNA and 5-aza-2′-deoxycytidine (5-aza-CdR) treatment (Dessain et al, 2000). For methylation-specific PCR with bisulphite-modified DNA, genomic DNAs from cells were isolated using Qiagen Tissue/Blood Kit (Qiagen), and bisulphite-modified according to published methods (Dessain et al, 2000). Promoter regions of hTERT were amplified from the nonmodified and the modified DNA. PCR reaction products were electrophoresed in 2% agarose and amplified DNA fragments were directly visualised under UV illumination.
Cells were incubated in the culture medium with 10 μ M 5-aza-CdR for 24 h on days 3 and 5, and then harvested on day 7 for analysis. Total RNA was prepared using Trizol™ reagent (Invitrogen). Randomly primed cDNAs were reverse-transcribed and hTERT cDNA was amplified as described above.
Results
Correlation between the expression of endogenous hTERT and telomerase activity in NHOF and NHOK
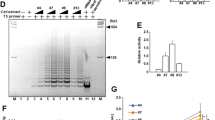
Our previous studies have shown the absence of telomerase activity in NHOF, its presence in exponentially replicating NHOK, and its gradual diminution during senescence in NHOK (Kang et al, 1998). This finding was confirmed in the present investigation, subjecting the exponentially replicating NHOF and NHOK, and senescent NHOK, to the telomeric repeat amplification (TRAP) assay (Figure 1A). Telomerase activity was not detected in replicating NHOF or in senescing NHOK, but was detected in exponentially proliferating NHOK (Figure 1).
(A) TRAP assay of NHOK and NHOF. Cellular extracts obtained after different PD were tested for telomerase activity. The positive control was 293 (adenovirus-transformed human embryonic kidney cells). The CHAPS buffer was used as a negative control. IC is an internal control amplification product provided in the assay kit. We repeated this experiment three times with NHOF and NHOK cultures derived from different donors and obtained similar results (data not shown). (B) Replicative lifespan of NHOK. The cell PD were calculated as described in Materials and Methods. NHOK at PD 15 and PD 20 were used for the TRAP assay.
The regulation of telomerase activity by expression of the hTERT gene was confirmed. The expression of full-length and splicing variants of hTERT was also determined using primers flanking the RT domain of hTERT (Ding et al, 2002). The full-length hTERT mRNA containing the functional RT domain was detected in exponentially replicating NHOK as a 457 bp band (Figure 2). The spliced variant of hTERT mRNA, also known as the β-deletion transcript containing a nonfunctional RT domain, was also identified in exponentially replicating NHOK as a 275 bp band (Figure 2). The β-deletion transcript causes a 183 bp deletion (bases 2342–2524) resulting in nonsense mutation, truncating the hTERT protein before the conserved RT motifs (Ulaner et al, 1998). hTERT mRNA was not detected in replicating NHOF or senescent NHOK which lacked telomerase activity (Figure 2). The presence of hTR mRNA in all tested cells indicated that hTR was not the limiting factor of the telomerase activity in NHOF and senescent NHOK (Figure 2).
Expression of hTERT and hTR mRNA in NHOK and NHOF. Three different NHOK cultures (NHOK-1, NHOK-2, and NHOK-3) and two different NHOF cultures (NHOF-1 and NHOF-2) from various donors were examined in this experiment. The level of hTERT mRNA was detected by RT–PCR and subsequent Southern blotting with 32P-labelled full-length hTERT cDNA. A full-length (457 bp) and spliced isoform (275 bp) of hTERT mRNA was detected in rapidly replicating NHOK. The level of hTR mRNA was determined by RT–PCR. β-Actin mRNA was amplified as a control.
Transcriptional activity of an exogenous hTERT promoter in NHOF and NHOK
The absence of hTERT mRNA in NHOF and senescent NHOK could result from either one of the following forms of negative control: (1) repressive alteration in the cis-acting hTERT promoter region or (2) defect in trans-acting regulatory factors required for promoter activity. To distinguish between the two possibilities, we examined the transcriptional activity of an exogenous hTERT promoter in NHOF and NHOK. SCC-4, a telomerase-positive human oral cancer cell line, served as positive control (Kang et al, 1998). The pGL3B-TRTP luciferase reporter plasmid containing a full-length hTERT promoter (nucleotides −1665 to +5) was transiently transfected into cells. The luciferase activity of pGL3B-TRTP was measured and compared with that of the pGL3-control plasmid, which was driven by the SV40 enhancer/promoter and used to monitor the transfection efficiency (Horikawa et al, 1999; Takakura et al, 1999).
As shown in Table 1, the pGL3B-TRTP plasmid expressed luciferase activity, that is, 9.8% of the pGL3-control, in the telomerase-positive human oral cancer cell line SCC-4. The same level of activity was found in exponentially replicating (PD 10) and senescing (PD 20) NHOK (7.0 and 6.9%), respectively. A similar level of activity (10.9% of the pGL3-control) was detected in hTERT-negative NHOF. These findings indicate that the necessary factors for hTERT expression were present and active in NHOF and senescent NHOK. It also suggests that negative alteration of the hTERT promoter is involved in repression of endogenous hTERT expression.
Methylation of the hTERT promoter in NHOF and senescent NHOK
Previous studies have shown that (1) promoter hypermethylation is frequently associated with gene silencing (Herman et al, 1999, 2) the telomerase gene has a typical CpG island domain for methylation in its promoter (Horikawa et al, 1999; Takakura et al, 1999), and (3) the human hTERT gene promoter is hypermethylated in cells with reduced telomerase activity (Devereux et al, 1999; Bechter et al, 2002). These findings suggested that the absence of hTERT gene expression in NHOF and senescent NHOK might be due to hypermethylation of the endogenous hTERT promoter. In the hTERT promoter, two CpG islands, the transcription start site and regions 3′ to the translation start site, have been well characterised (Cong et al, 1999; Horikawa et al, 1999; Takakura et al, 1999; Dessain et al, 2000). Therefore, it was possible to determine the presence of methylation in both CpG islands in NHOF and senescent NHOK by using bisulphite treatment of DNA and subsequent methylation-specific PCR (Dessain et al, 2000).
In NHOF, as shown in Figure 3, the region 3′ to the translation start site (TL) was completely methylated, while the region 3′ to the transcription start site (TC) was not methylated (Figure 3A). In rapidly proliferating NHOK (PD 11) that expressed hTERT mRNA and telomerase activity, both CpG islands in the hTERT promoter were hypomethylated (Figure 3B). Senescing NHOK (PD 20) showed methylation in both CpG islands (Figure 3C). With a reduced number of PCR cycles (15 cycles), the fraction of cells containing the methylated promoter was greater than that of cells containing the unmethylated promoter (Figure 3C). The methylation status of the hTERT promoter in the telomerase-positive cell line SCC-4 was also examined. Both CpG islands in the hTERT promoter were not methylated in the cell line (Figure 3D).
Methylation-specific PCR of the hTERT CpG islands in NHOF and NHOK. Genomic DNA was treated with bisulphite and tested with transcription start site (TC) and translation start site (TS) primer sets specific for either unmethylated (U) or methylated (M) hTERT promoter. (A) Methylation status of hTERT promoter in rapidly replicating NHOF (PD 18). The promoter was amplified with 25 cycles. (B) Methylation status of hTERT promoter in rapidly replicating NHOK (PD 15). The promoter was amplified with 25 cycles. (C) Methylation status of hTERT promoter in senescent NHOK (PD 20). The promoter was amplified with 15 or 25 cycles. (D) Methylation status of hTERT promoter in SCC-4. The promoter was amplified with 25 cycles.
Introduction of endogenous hTERT expression in NHOF and senescing NHOK exposed to 5-aza-CdR
We investigated whether hypermethylation was solely responsible for repression of the hTERT gene expression by exposing the cells to 5-aza-CdR, the potent demethylating agent (Bender et al, 1998). The concentrations of 5-aza-CdR utilised in the present study were based on the studies reported by others (Devereux et al, 1999; Dessain et al, 2000). Furthermore, we determined the cytotoxic effect of 5-aza-CdR (at a range of 1–10 μ M) in exponentially proliferating NHOF and NHOK for up to 3, 7, or 10 days, and found that 1 or 5 μ M of 5-aza-CdR did not demonstrate any cytotoxic effect in these cells during the 7-day period, by determining the proliferation rate of cells as described elsewhere (Kang et al, 2000). NHOF and senescing NHOK were cultured in the absence or presence of 5 μ M 5-aza-CdR for 3, 5, and 7 days, and hTERT gene transcription was tested by RT–PCR Southern blotting (Figure 4).
(A) Re-expression of endogenous hTERT mRNA in rapidly replicating NHOF (PD 24) exposed to 5-aza-CdR. (B) Re-expression of endogenous hTERT mRNA in senescent NHOK (PD 19) exposed to 5-aza-CdR. The cells were treated with 5 μ M of 5-aza-CdR for 3, 5, and 7 days and analysed for hTERT mRNA expression using RT–PCR Southern blotting. β-Actin mRNA was amplified as a control.
After 5 days of treatment, NHOF expressed a full-length (457 bp) and a spliced isoform (275 bp) of hTERT mRNA, whose expression was maintained during the entire 7-day treatment (Figure 4A). Senescing NHOK after 3 days of treatment with 5-aza-CdR re-expressed a full-length (457 bp) and a spliced variant (275 bp) of hTERT mRNA (Figure 4B). With longer exposure of the gel, the full-length hTERT mRNA was clearly visible in the 5-aza-CdR-treated NHOK (data not shown). Telomerase activity tested by the TRAP assay was not detected in NHOF and senescing NHOK exposed to 5-aza-CdR (data not shown). These results suggest that hTERT promoter hypermethylation in NHOF and senescent NHOK is linked to gene silencing, which can be reversed by demethylation.
Discussion
Activation of telomerase activity is thought to be a critical step in cellular immortalisation and transformation. Since telomerase activity is primarily regulated through hTERT expression, understanding of hTERT regulation in normal cells is crucial for the understanding of carcinogenesis. Our present studies provide evidence that methylation of the hTERT promoter is responsible for the repression of hTERT expression, which results in the lack of telomerase activity in NHOF and senescent NHOK.
Transcriptional activation has been considered a major mechanism for the regulation of the hTERT gene (Poole et al, 2001). Consistent with previous observations (Kim et al, 1994; Shay and Bacchetti, 1997), we found that expression of hTERT is a limiting factor for telomerase activity in NHOF and NHOK. Also, the presence of hTERT mRNA encoding functional or defective spliced isoforms has been demonstrated in several types of normal and pathogenic human cells (Ulaner et al, 1998, 2000; Liu et al, 1999). The splicing pattern of hTERT mRNA has been considered part of a regulatory mechanism for telomerase activity, but the details are unknown (Kilian et al, 1997; Ulaner et al, 1998, 2000; Wick et al, 1999). The β-deletion transcript causes premature termination of translation, resulting in truncation of the protein, and has been observed in various cell types (Nakamura et al, 1997; Wick et al, 1999; Ding et al, 2002). In the present study, we have detected the full-length and the β-deletion transcripts of hTERT in normal oral keratinocytes. Similar to our observations, coexpression of the full-length and the β-deletion transcripts have been reported in normal ovarian tissues (Ulaner et al, 2000) and human papillomavirus-immortalised endocervical cells (Ding et al, 2002). According to a previous report (Yi et al, 2001), a β-deletion splice variant was dominantly expressed (80–90% of total hTERT mRNA) in various human epithelial cells. Since NHOK is an epithelial cell, the domination of the β-deletion splice variant is not surprising. Although the reason for the dominant reactivation of the β-deletion splice variant upon the 5-aza-CdR treatment is not known, we could not rule out the possibility that the half-life (stability) of the β-deletion splice variant is longer than the full-length hTERT mRNA, and autosecretion of TGF-β retains high expression of the inactive β-deletion splice variant in NHOK culture (Cerezo et al, 2002).
After testing the transcriptional activity of an exogenous hTERT promoter in NHOF, rapidly proliferating NHOK and senescent NHOK, we found significant transcriptional activity in all tested cells, regardless of the presence of telomerase activity. The promoter region (nt –1665 to +5) examined in this study included the 59 bp region (nt −208 to −150) that was responsible for the maximal promoter activity (Horikawa et al, 1999). The data showed that in NHOF and senescing NHOK: (1) the transcriptional machinery necessary for hTERT promoter activity in replicating NHOF functioned as well as in NHOK-expressing hTERT mRNA and (2) the transcriptional machinery necessary for the hTERT promoter activity was not activated or inactivated during senescence in NHOK. Therefore, the absence of hTERT expression does not appear to be caused by a defective transcriptional machinery, but by an altered endogenous hTERT promoter.
In contrast with other reports of unmethylated hTERT promoters in normal fibroblasts (Devereux et al, 1999; Dessain et al, 2000), our studies show that hTERT expression appears to be correlated with the methylation status of the promoter in NHOF, suggesting that promoter methylation does provide a means to regulate hTERT expression in cells. To rule out the possibility of experimental errors and donor variation, we performed two assays in three strains of NHOF from different donors and obtained consistent reproducible results. Therefore, the hTERT promoter is tissue-specifically methylated and silenced in normal human fibroblasts (Imamura et al, 2001; Shiota et al, 2002).
Previously, we reported that replication arrest preceded differentiation in NHOK serially subcultured in vitro (Kang et al, 2000). The silencing of hTERT during squamous cell differentiation was demonstrated (Harle-Bachor and Boukamp, 1996). Based on these observations, we speculate that the silencing of hTERT due to promoter methylation is also in part associated with the silencing of hTERT during squamous differentiation in vivo.
Hypermethylation of the hTERT promoter during senescence in normal oral keratinocytes was correlated with the diminution of telomerase activity and hTERT mRNA expression. The involvement of promoter methylation in hTERT gene regulation during senescence in oral keratinocytes was further confirmed by induction of hTERT expression in senescent NHOK exposed to 5-aza-CdR. This finding supported the concept that the differentiation of cells is associated with changing methylation status at many loci in the genome (Shiota et al, 2002). While hTERT expression was restored by exposure to 5-aza-CdR, telomerase activity was not. Although the reason for inability of the chemical to restore telomerase activity is not known, we speculate that the inhibitory effect of 5-aza-CdR on DNA synthesis and telomerase activity may be the cause. 5-aza-CdR inhibited both DNA synthesis and telomerase activity in exponentially replicating NHOK (our unpublished data).
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 72: 141–196
Bechter OE, Eisterer W, Dlaska M, Kuhr T, Thaler J (2002) CpG island methylation of the hTERT promoter is associated with lower telomerase activity in B-cell lymphocytic leukemia. Exp Hematol 30: 26–33
Bender CM, Pao MM, Jones PA (1998) Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res 58: 95–101
Cerezo A, Kalthoff H, Schuermann M, Schafer B, Boukamp P (2002) Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J Cell Sci 115: 1305–1312
Cong YS, Wen J, Bacchetti S (1999) The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet 8: 137–142
Dessain SK, Yu H-Y, Reddel RR, Beijersbergen RL, Weinberg RA (2000) Methylation of the human telomerase gene CpG island. Cancer Res 60: 537–541
Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC (1999) DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res 59: 6087–6090
Ding Z, Green AG, Yang X, Chernenko G, Tang SC, Pater A (2002) Retinoic acid inhibits telomerase activity and downregulates expression but does not affect splicing of hTERT: correlation with cell growth rate inhibition in an in vitro cervical carcinogenesis/multidrug-resistance model. Exp Cell Res 272: 185–191
Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu F, Le S, West MD, Harley CB, Andrews W, Greider CW, Villeponteau B (1995) The RNA component of human telomerase. Science 269: 1236–1241
Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J (2002) Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer 101: 335–341
Harle-Bachor C, Boukamp P (1996) Telomerase activity in the regenerative basal layer of the epidermis inhuman skin and in immortal and carcinoma-derived skin keratinocytes. Proc Natl Acad Sci USA 93: 6476–6481
Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO (1997) A mammalian telomerase-associated protein. Science 275: 973–977
Herman JG (1999) Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol 9: 359–367
Horikawa I, Cable PL, Afshari C, Barrett JC (1999) Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res 59: 826–830
Imamura T, Ohgane J, Ito S, Ogawa T, Hattori N, Tanaka S, Shiota K (2001) CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. Genomics 76: 117–125
Kang MK, Bibb C, Baluda MA, Rey O, Park N-H (2000) In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res 258: 288–297
Kang MK, Guo W, Park N-H (1998) Replicative senescence of normal human oral keratinocytes is associated with the loss of telomerase activity without shortening of telomeres. Cell Growth Differ 9: 85–95
Kang MK, Swee J, Kim RH, Baluda MA, Park NH (2002) The telomeric length and heterogeneity decrease with age in normal human oral keratinocytes. Mech Ageing Dev 123: 585–592
Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 6: 2011–2019
Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015
Liu K, Schoonmaker MM, Levine BL, June CH, Hodes RJ, Weng NP (1999) Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc Natl Acad Sci USA 96: 5147–5152
Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90: 785–795
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959
Poole JC, Andrews LG., Tollefsbol TO (2001) Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT). Gene 269: 1–12
Shay J, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791
Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, Tanaka S, Hattori N (2002) Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 7: 961–969
Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M (1999) Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res 59: 551–557
Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR (1998) Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res 58: 4168–4172
Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR (2000) Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int J Cancer 85: 330–335
Wick M, Zubov D, Hagen G (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232: 97–106
Yi X, Shay JW, Wright WE (2001) Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 29: 4818–4825
Zingg JM, Jones PA (1997) Genetic and epigenetic aspects of DNA methylation on genome expression, evolution, mutation and carcinogenesis. Carcinogenesis 18: 869–882
Acknowledgements
We thank Dr JC Barrett (NIEHS) for providing the pGL3B-TRTP plasmids. This work was supported in part by the grants (DE14147 and DE14635) funded by the National Institute of Dental and Craniofacial Research (NIDCR).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Shin, KH., Kang, M., Dicterow, E. et al. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer 89, 1473–1478 (2003). https://doi.org/10.1038/sj.bjc.6601291
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6601291
Keywords
This article is cited by
-
RIF1 promotes human epithelial ovarian cancer growth and progression via activating human telomerase reverse transcriptase expression
Journal of Experimental & Clinical Cancer Research (2018)
-
Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer
Journal of Biomedical Science (2018)
-
Maternal telomere length inheritance in the king penguin
Heredity (2015)
-
Genetic and epigenetic trends in telomere research: a novel way in immunoepigenetics
Cellular and Molecular Life Sciences (2015)
-
Ferrocenylnaphthalene Diimide-Based Electrochemical Detection of Aberrant Methylation in hTERT Gene
Applied Biochemistry and Biotechnology (2014)