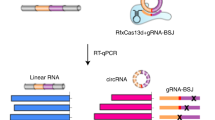
Abstract
Circular RNAs (circRNAs) produced from back-spliced exons are widely expressed, but individual circRNA functions remain poorly understood owing to the lack of adequate methods for distinguishing circRNAs from cognate messenger RNAs with overlapping exons. Here, we report that CRISPR–RfxCas13d can effectively discriminate circRNAs from mRNAs by using guide RNAs targeting sequences spanning back-splicing junction (BSJ) sites featured in RNA circles. Using a lentiviral library that targets sequences across BSJ sites of highly expressed human circRNAs, we show that a group of circRNAs are important for cell growth mostly in a cell-type-specific manner and that a common oncogenic circRNA, circFAM120A, promotes cell proliferation by preventing the mRNA for family with sequence similarity 120A (FAM120A) from binding the translation inhibitor IGF2BP2. Further application of RfxCas13d–BSJ-gRNA screening has uncovered circMan1a2, which has regulatory potential in mouse embryo preimplantation development. Together, these results establish CRISPR–RfxCas13d as a useful tool for the discovery and functional study of circRNAs at both individual and large-scale levels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All sequencing datasets have been deposited in NCBI GEO (GSE149690, GSE149691, and GSE149692) and National Omics Data Encyclopedia (OEP000887, OEP000888 and OEP000889). The total RNA sequences of human H9, FB and PA1 cell lines were downloaded from NCBI GEO (GSE73325). RNA-seq datasets of SH-SY5Y were downloaded from NCBI GEO (GSE65926). RNA-seq datasets of HepG2, K562 cell lines and 11 different human tissues were downloaded from ENCODE Project Consortium (https://www.encodeproject.org/). AGO2 PAR-CLIP and miRNA-seq datasets of HEK293 cells were downloaded from NCBI GEO (GSE43573 and GSE58127). IGF2BP2 of eCLIP–seq datasets in K562 cells was downloaded from NCBI GEO (GSE91445). The single-cell RNA-seq transcriptome datasets in mouse preimplantation embryos were from NCBI GEO (GSE53386). Source data are provided with this paper.
Code availability
The custom Perl and Shell scripts for the computational pipeline of Cas13d-mediated circRNA screen (CDCscreen) to identify negatively selected functional circular RNAs in this paper is available at https://github.com/YangLab/CDCscreen.
References
Li, X., Yang, L. & Chen, L. L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442 (2018).
Chen, S. et al. Widespread and functional RNA circularization in localized prostate cancer. Cell 176, 831–843(2019).
Guarnerio, J. et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 166, 1055–1056 (2016).
Legnini, I. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22 (2017).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
Xia, P. et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701 e687 (2018).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell 60, 385–397 (2015).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676 e614 (2018).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Yan, W. X. et al. Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327 (2018).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Chen, G. W., Shi, Y. T., Zhang, Y. & Sun, J. Y. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco. Targets Ther. 10, 5783–5794 (2017).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880 e821 (2019).
Zhang, X. O. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26, 1277–1287 (2016).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 (2017).
Zheng, Q. P. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 (2016).
Lee, M., Kim, E. J. & Jeon, M. J. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget 7, 8726–8742 (2016).
Mu, Y. C. et al. NUPR1 maintains autolysosomal efflux by activating SNAP25 transcription in cancer cells. Autophagy 14, 654–670 (2018).
Vincent, E. E. et al. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 (2015).
Wallin, J. J. et al. Nuclear phospho-Akt increase predicts synergy of PI3K inhibition and doxorubicin in breast and ovarian cancer. Sci. Transl. Med. 2, 48ra66 (2010).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Bartolome, R. A. et al. IL13 receptor α2 signaling requires a scaffold protein, FAM120A, to activate the FAK and PI3K pathways in colon cancer metastasis. Cancer Res 75, 2434–2444 (2015).
Li, Z. et al. An HMGA2–IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev. Cell 23, 1176–1188 (2012).
Dai, N. et al. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab. 21, 609–621 (2015).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Zhou, C. et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262–2276 (2017).
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Bultman, S. et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 (2000).
Bultman, S. J. et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 20, 1744–1754 (2006).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Yin, Q. F. et al. Long noncoding RNAs with snoRNA ends. Mol. Cell 48, 219–230 (2012).
Li, S. et al. Screening for circRNAs with functional potential using the RfxCas13d/gRNA library. Protocol Exchange https://doi.org/10.21203/rs.3.pex-1181/v1 (2020).
Yang, L., Duff, M. O., Graveley, B. R., Carmichael, G. G. & Chen, L. L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 12, R16 (2011).
Wang, Y. et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 20, 1145 (2018).
Dong, R., Ma, X. K., Li, G. W. & Yang, L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteom. Bioinforma. 16, 226–233 (2018).
Acknowledgements
We thank Chen and Yang laboratories for discussion. This work was supported by the Chinese Academy of Sciences (CAS) (XDB19020104), the National Natural Science Foundation of China (NSFC) (91940303, 31821004, 31725009) and the HHMI International Program (55008728) to L.-L.C. NSFC (31730111, 31925011, 91940306) to L.Y.; NSFC (31730062, 31821004) and the Shanghai Municipal Commission for Science and Technology (19411951800) to J.L.; NSFC (31801073) and the Youth Innovation Promotion Association CAS to W. X.
Author information
Authors and Affiliations
Contributions
L.-L.C. supervised and conceived the project. L.-L.C., L.Y., J.L., S.L., X.L., W.X. and L.Z. designed experiments. L.Z. performed circRNA screening in mouse embryos, supervised by J.L.; S.L., X.L., L.-Z.Y., S.-M.C., C.-X.L., S.-K.G., L.S., M.W., X.T., J.-L.Z., X.G., J.Z. and J.W. performed all other experiments; W.X., Y.-N.L. and L.Y. preformed computational analyses. L.-L.C. and L.Y. wrote the paper with input from S.L., X.L., W.X and J.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Lei Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
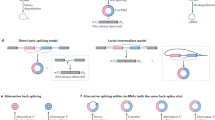
Extended Data Fig. 1 Evaluation of RfxCas13d knockdown efficiency and specificity.
a, Strategies of circRNA knockdown (KD) by RNAi, shRNA and ASO. b, Strategies of circRNA knockout by CRISPR/Cas9. Top, genome editing is used to remove circRNA-formed exons5; bottom, genome editing destroys the intronic RNA pair to block circRNA expression6,7. c, Schematic of circRNA KD by Cas13. Three gRNAs targeting the back-splicing junction site (BSJ, red arrow) were designed for each circRNA (BSJ-gRNAs). Expression of circRNAs and cognate linear mRNAs were detected by qRT–PCR. d,e, Expression of Cas13 proteins were detected by msfGFP fluorescence (d) and WB (e) in 293FT cells after 48 h transfection. Scale bar, 200 μm. Results are representative of two independent experiments. f, Comparison of different Cas13 proteins-mediated KRAS knockdown efficiencies with two position-matched guides revealed that RfxCas13d is the best effector14. g, Evaluation of different Cas13-protein-mediated KD on circRNAs. n = 63 biologically independent samples. Data are presented as means values ± s.d. h, NB confirmed the circPOLR2A knockdown by RfxCas13d/BSJ-gRNAs. Results are representative of two independent experiments. i,j, BSJ gRNAs (i) or RfxCas13d (j) alone did not affect circPOLR2A and circRTN4 expression. k, gRNAs with partial sequences replaced by adjacent linear exon (gRNA-L) led to linear but not circular RNA KD. Top, schematic of gRNA-L. Bottom, KD efficiencies of circPOLR2A and circRTN4 as well as their corresponding linear RNAs. l, Replacement of a 10nt (scram/mismatch) on either side of an originally highly effective 30nt gRNA (blue bars) completely blocked the KD effect on the target mRNAs. Two 30nt effective gRNAs and the corresponding 10(scram/mismatch) + 20nt gRNAs on two individual mRNAs were tested. (f,i,j,k,l) All expression levels of RNAs were detected by qRT-PCR and were normalized to ACTB, means ± s.d. were from three independent experiments. (f,g,j,k,l) *: P < 0.05; **: P < 0.01; ***: P < 0.001; ns, not significant, two-tailed student’s t-test.
Extended Data Fig. 2 RfxCas13d is the best effector to mediate circRNA knockdown.
a,b, Evaluation of Cas13 proteins for knocking down circRNAs (circPOLR2A and circRTN4) and their cognate mRNAs in HeLa cells revealed that RfxCas13d is the best effector for circRNA-specific KD. b, n = 63 biologically independent samples and transcript levels were normalized to ACTB. Data are presented as means values ± s.d. c, RfxCas13d/BSJ-gRNAs specifically and robustly knocked down circRNAs, but not their cognate mRNAs in HeLa cells. d, Mismatch tolerance of RfxCas13d for circRNA targeting. Guides containing single or double mismatches at varying positions across spacer sequences for circPOLR2A are shown in purple; bases flanking the BSJ site are shown in red. e, Efficiency and specificity of RfxCas13d for circRNA and cognate linear mRNA KD. Top, schematic of gRNAs targeting circPVT1 tiled 10-nt increments away from the BSJ site. Bottom, KD efficiencies of circRNA and linear RNA by each gRNA and RfxCas13d. f-g, Arrayed KD screen of 23 guides evenly tilled across BSJ of circHIPK3 (f) and circRTN4 (g). Position-effect of BSJ-gRNAs for RfxCas13d-mediated KD of circHIPK3 (f) and circRTN4 (g) at the single nucleotide level. Twenty-three guides tiled across the BSJ of circHIPK3 (f) and circRTN4 (g) are listed. KD efficiencies of circHIPK3 (f) or circRTN4 (g) and linear HIPK3 (f) or linear RTN4 (g) by each BSJ-gRNA were shown in the right. (a,c,d,e,f,g) All expression levels of RNAs were detected by qRT-PCR and were normalized to ACTB, means ± s.d. were from three independent experiments. (a,b,c) **: P < 0.01; ***: P < 0.001; ns, not significant, two-tailed student’s t-test.
Extended Data Fig. 3 Characterization of the BSJ-gRNA library targeting 762 circRNAs.
a, Calculation of circRNA copy numbers (using circPOLR2A as an example) in H9, HT29, HeLa and 293FT cells. According to the FPBcirc of circPOLR2A and other circRNAs in H9, HT29, HeLa and 293FT cells, and the six copies of circPOLR2A per HeLa cell17, we calculated the copy number of all circRNAs, respectively. Copies of circPOLR2A per cell were listed. b, Schematic of circRNA copy number calculation in H9, HT29, HeLa and 293FT cells. c, Representation of 762 candidate circRNAs designed with BSJ-gRNAs in the library in different cell lines. d, Matched length distribution of 762 candidate circRNAs (solid line) and total circRNAs (dashed line). Density curve and pie chart show that more than 80% of 762 candidate circRNAs and total circRNAs are less than 1,000nt. e, Cumulative distribution of the number of reads per gRNA of constructed libraries. The red line indicates that less than 0.2% of gRNAs are covered by less than 100 reads. f, WB confirmed the stable expression of Flag-RfxCas13d in HT29, 293FT and HeLa cells used for screening. Results are representative of two independent experiments. g, KD efficiency of circRNAs remained unchanged in 30 days. RfxCas13d/BSJ-gRNA infected HT29, 293FT and HeLa cells were collected at a series of timepoints (day 1, 10, 20, 30). KD efficiency of different circRNAs was detected by qRT-PCR and were normalized to ACTB. Means ± s.d. were from three independent experiments. h, Pearson correlation coefficient (PCC) between replicates (rep) of D1 and D30 samples in HT29, 293FT and HeLa cells. Two biologically independent experiments were performed at D1 and D30 in each cell line. i, Scatter plot of fold change of paired controls and circRNA BSJ-gRNAs between D1 and D30 samples in HT29, 293FT and HeLa cells. The grey dashed lines indicate 2 or 0.5 fold change, respectively.
Extended Data Fig. 4 Pipelines used to identify negatively selected candidate circRNAs important for cell growth.
a, Analysis pipeline to identify negatively selected circRNA candidates by CDCscreen. Uniquely-mapped reads of each gRNA were normalized, and averaged normalized reads of two biological replicates were used for subsequent analyses. P value of negatively selected circRNA was computed by the permutation test of MAGeCK, and mean fold change of negatively-selected gRNAs targeting the same circRNAs between D30 and D1 treatments was obtained from normalized gRNA reads. Only expressed circRNAs with FPBcirc > 0 were calculated. Finally, circRNAs with CDCscreen score ≥ 2, ≥ 2 negatively-selected gRNAs with FC ≤ 0.667 were identified as circRNA candidates to promote cell proliferation. b, Rank of negatively selected candidate circRNAs by CDCscreen scores in HT29, 293FT and HeLa cells. CircRNAs with CDCscreen score ≥ 2 and with ≥ negatively-selected gRNAs and FC ≤ 0.667 were sub-grouped by red dashed line. Examples of candidate circRNAs were marked in each cell line. c, Each item in the table corresponds to each step of calculation of candidate circRNAs in CDCscreen pipeline shown in (a). CircFAM120A is shown as an example, and related analyses of all other circRNAs are listed in Supplementary Table 4. d, Analysis of gRNAs in FC ≤ 0.667 that identifies the same circRNA as a target gene. The number of circRNA candidates, the average total gRNAs and altered gRNAs in FC ≤ 0.667 (purple) in each cell line are shown. n = 67 circRNAs in HT29, n = 62 circRNAs in 293FT, n = 63 circRNAs in HeLa. Data are presented as means values ± s.d. e, False discovery rate (FDR) of circRNA candidates with CDCscreen score ≥ 2 is less than 0.1 for in vitro (Fig. 2b) and in vivo (Extended Data Fig. 7a) screens.
Extended Data Fig. 5 Validation of candidate circRNAs by the RfxCas13d/BSJ-gRNA system in cell proliferation.
a, Heatmap display of the relative KD efficiency, cell proliferation and fold change of candidate circRNAs in HT29 (purple), 293FT (orange) and HeLa (green) cells with single BSJ-gRNA that targets each candidate circRNA. b,c, KD of circKLHL8 by RfxCas13d inhibited cell proliferation in 293FT (b) and HeLa cells (c), as revealed by MTT cell proliferation assay (middle panels) and cell confluency calculated by the surface area occupied by cells (right panels); KD efficiencies were showed on left panels. d, KD of circHIPK3 by RfxCas13d inhibited cell proliferation in HeLa cells, as revealed by MTT cell proliferation assay (middle) and by cell confluency calculated by the surface area occupied by cells (right); KD efficiencies were showed on left. e, KD of circKLHL8 by shRNAs inhibited cell proliferation in 293FT cells. KD efficiencies were showed on left; MTT cell proliferation assays were shown on right. f, KD of circKLHL8 and circHIPK3 by shRNAs in HeLa cells. g, KD of circKLHL8 or circHIPK3 by shRNAs inhibited cell proliferation in HeLa cells, as revealed by MTT cell proliferation assays. (a,b,c,d,e,f) All expression levels of RNAs were detected by qRT-PCR and were normalized to ACTB. Means ± s.d. were from three independent experiments. *: P < 0.05; **: P < 0.01; ns, not significant, two-tailed student’s t-test.
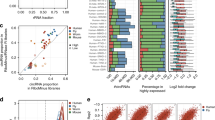
Extended Data Fig. 6 Distribution of CDCscreen scores and expression levels of circRNAs that have cell type-specific effects on cell growth.
a, Distribution of CDCscreen scores of circRNAs with cell-type specific effects on cell proliferation in HT29 (n = 59 circRNAs), 293FT (n = 47 circRNAs) and HeLa (n = 53 circRNAs) cell, respectively. See ‘Data visualization’ in the Methods for definitions of box plot elements. b, Expression (shown by FPBcirc, log10) of circRNAs shown in (a) in HT29 (n = 59 circRNAs), 293FT (n = 47 circRNAs) and HeLa (n = 53 circRNAs) cells. See ‘Data visualization’ in the Methods for definitions of box plot elements. c. Expression (shown by FPBcirc, log10) of validated circRNAs (n = 6) in Fig. 2d that have cell-type specific effects on cell proliferation in HT29, 293FT and HeLa cells. See ‘Data visualization’ in the Methods for definitions of box plot elements. d, CircFAM120A KD by RfxCas13d/BSJ-gRNAs inhibited HT29, 293FT or HeLa cell proliferation, as revealed by cell confluency assays. e, CircFAM120A KD by shRNAs in HeLa cells. Expression of circRNAs and cognate linear RNAs was detected by qRT-PCR and was normalized to ACTB. f, CircFAM120A KD by shRNAs inhibited cell proliferation in HeLa cells, as revealed by MTT assays. g, CircFAM120A KD by shRNAs in 293FT cells. Expression of circRNAs and cognate linear RNAs was detected by qRT-PCR and was normalized to ACTB. h, CircFAM120A KD by shRNAs inhibited cell proliferation in 293FT cells, as revealed by MTT assays. (d,e,f,g,h) Means ± s.d. were from three independent experiments. (a,b,d,e,f,g,h) *: P < 0.05; **: P < 0.01; ***: P < 0.001; ns, not significant, two-tailed student’s t-test.
Extended Data Fig. 7 Overview of sequencing analyses of in vivo screens using BSJ-gRNA libraries targeting 2,908 circRNAs.
a, Construction of the gRNA library targeting 2,908 circRNAs. One paired control gRNA (n = 2,908) and three BSJ-gRNAs (circRNA gRNAs, n = 8,724) were designed for each candidate circRNA. b, Cumulative distribution of the number of reads per gRNA of constructed libraries. The red line indicates that less than 1% of gRNAs are covered by less than 100 reads. c, Representation of 2,908 candidate circRNAs in different human tissues41 constructed in the library. On average, over 90% of top 100 abundant circRNAs in each tissue were included in the list of 2,908 candidate circRNAs. d, In vivo screen of circRNAs important for cell growth and proliferation. The gRNA lentiviral library was individually delivered into HT29 cells stably expressing RfxCas13d. Infected cells were enriched after 7 days and injected subcutaneously to nude mouse for 22 days. Genomic DNAs from infected cells were extracted at day 1 (D1) and 30 (D30) for gRNA amplification and deep sequencing. e, The Pearson correlation coefficient (PCC) between replicates (rep) of D1 and D30 in vivo samples in HT29. Two biologically independent experiments were performed at D1 and D30. f, Scatter plot of fold change of paired controls and circRNA BSJ-gRNAs between D1 and D30 in vivo samples in HT29. The grey dashed lines indicate 2 or 0.5 fold change, respectively. g, Analysis of gRNAs in FC ≤ 0.5 that identifies the same circRNA as a target gene. The number of circRNA candidates (n = 79), the average total gRNAs and altered gRNAs in FC ≤ 0.5 (purple) in in vivo screen are shown. Data are presented as means values ± s.d.
Extended Data Fig. 8 RNA-seq analysis of RfxCas13d/BSJ-gRNA- or shRNA- mediated circFAM120A KD.
a, Mapping statistics of two biological replicates of the poly(A) + RNA-seq datasets in 293FT cells with RfxCas13d/BSJ-gRNA- or shRNA- mediated circFAM120A KD. b, Heatmap of Spearman’s rank correlation coefficient for log2(FPKM) values of all linear mRNAs detected in RNA-seq libraries between targeting and non-targeting replicates for RfxCas13d/BSJ-gRNA- or shRNA- mediated circFAM120A KD. c, Expression levels in log2(FPKM) values of all genes detected in RNA-seq libraries of non-targeting control (x-axis) compared to circFAM120A-targeting conditions (y-axis) by RfxCas13d/BSJ-gRNA (red) or shRNA (blue). Means of two biological replicates were shown. d, A workflow shows the selection of candidate genes after circFAM120A KD by RfxCas13d/BSJ-gRNA. e, Heatmap of DEGs (n = 97) detected after circFAM120A KD by RfxCas13d/BSJ-gRNA. f, Enrichment of DEGs from RNA-seq after circFAM120A KD by RfxCas13d/BSJ-gRNA. The x axis shows the ratio of the number of genes in a given category of functional annotations divided by the total number of DEGs. The y axis shows categories of functional annotations. P values were calculated based on the Fisher’s exact test. g, Validation of DEGs associated with cell proliferation after circFAM120A KD by RfxCas13d/BSJ-gRNA in 293FT cells. All transcripts were normalized to ACTB, n = 3 independent experiments. ***: P < 0.001, two-tailed student’s t-test. h, Cytoplasmic distribution of circFAM120A. i, Prediction of AGO2-binding peaks in circFAM120A. Top, genomics locus and diagram of linear FAM120A and circFAM120A (shown as magenta cylinders). Blue and magenta arrows indicate location of primer for linear FAM120A or circFAM120A. Bottom, predicted miRNA target sites by TargetScan and AGO2 binding peaks from PAR-CLIP data in HEK293FT cells (GEO: GSE43573). j, A schematic to show predication of potential circRNA-miRNA target sites. k, CircFAM120A did not interact with AGO2 by RIP in 293FT cells using anti-AGO2 antibodies. (g,h,k) All RNA levels were detected by qRT-PCR and means ± s.d. were from three independent experiments.
Extended Data Fig. 9 CircFAM120A promotes cell proliferation by regulating its parental gene translation in an IGF2BP2-dependent manner.
a,c, KD of FAM120A mRNA by shRNAs in 293FT (a) and HT29 (c) cells. b, Stable KD FAM120A by two shRNAs in 293FT cells, confirmed by one WB experiment. d, KD of FAM120A mRNA by shRNAs inhibited cell proliferation in HT29 cells, revealed by MTT assays. e, circFAM120A KD by two gRNAs reduced FAM120A protein expression in HT29 cells, confirmed by WB of one experiment; see also three independent experiments in 293FT cells (Fig. 3c). f, Schematic of sucrose gradients used to segregate fractions in polysome profiling assays. The light fraction contains ribosome subunits and single ribosomes; the heavy fraction contains polyribosomes. g, CircFAM120A KD led to altered distribution of FAM120A mRNA on ribosomes. FAM120A mRNA in individual fractions were measured by qRT-PCR and were normalized to GAPDH. h, Stability of FAM120A protein remains unchanged after circFAM120A KD. Cycloheximide (CHX) was used to inhibit translation. Quantification of results from duplicated assays was shown underneath. i, Absolute quantification of IGF2BP2 copies per cell. Results are representative of three experiments. j, Copies of circFAM120A, linear FAM120A and IGF2BP2 protein per HT29, 293FT or HeLa cells. k, IGF2BP2 was mainly localized in the cytoplasm. Left, representative image of IGF2BP2 immunofluorescence. Right, statistics of IGF2BP2 signals in each image. n = 14 images. Data are presented as means values ± s.d. l, Absolute copies of circFAM120A and linear FAM120A associated with IGF2BP2, calculated from IGF2BP2 RIP assays in 293FT cells (Fig. 3g). m-n, Distribution of IGF2BP2 protein, circFAM120A and FAM120A mRNA in fractions of polysome profiling of 293FT cells. o, m6A promoted circFAM120A binding to IGF2BP2 in vitro, detected by NB. p, m6A enhanced the capability of circFAM120A to compete with linear FAM120A binding to IGF2BP2 in vitro. Dig-labeled linear FAM120A associated with His-IGF2BP2 was detected by NB. q, Expression of FAM120A protein was partially rescued by loss of IGF2BP2 in circFAM120A KD 293FT cells, revealed by WB. (a,c,d,g,h,m,n,o,p,q) Means ± s.d. are from three independent experiments. (a,c,d,g,o,p,q) *: P < 0.05; **: P < 0.01, two-tailed student’s t-test. (h, o, p, q) Statistics were quantified from three independent experiments processed in parallel.
Extended Data Fig. 10 Application of RfxCas13d/gRNA to interfere RNA expression during mouse preimplantation development.
a, KD of Brg1 and Kras by RfxCas13d/gRNA in zygotes. Expression of mRNAs was detected by qRT-PCR and normalized to Actb. b, Representative images of reduced blastocyst formation 96 h after microinjection of RfxCas13d mRNA and the gRNA targeting Brg1 mRNA into mouse zygotes. An example of failed blastocyst formation is shown by red arrows and enlarged view. c, Representative images of embryogenesis after microinjection of RfxCas13d mRNA and the gRNA targeting Kras mRNA into mouse zygotes. No aberrant mouse preimplantation development was observed. d, Effect of Brg1 (b) and Kras (c) KD on blastocyst formation 96 h after microinjection of RfxCas13d mRNA and BSJ-gRNAs into mouse zygotes. e, Diagrams of mouse circMan1a2 and human circMAN1A2 are shown as magenta cylinders. f, KD of circMan1a2 in zygotes led to reduced blastocyst formation 72 h after microinjection of RfxCas13d mRNA and BSJ-gRNAs into mouse zygotes. Representative images of at 2-cell, 4-cell, morula and blastocyst stages are shown; an example under each condition is highlighted by red line and enlarged view. g, KD of circDcbld2 by RfxCas13d/BSJ-gRNA in zygotes. Expression of circDcbld2 was detected by qRT-PCR and normalized to Actb. h, Images of normal embryonic morphologies at 2-cell, 4-cell, morula and blastocyst stages under circDcbld2 KD by RfxCas13d/BSJ-gRNA in zygotes. i, Statistics of circDcbld2 KD effectd on morula and blastocyst formation after microinjection of RfxCas13d mRNA and BSJ-gRNAs into mouse zygotes. j,k, Implantation of RfxCas13d-gRNA (circMan1a2 KD)-injected zygotes to pseudo-pregnant female mice led to retarded mouse postimplantation development in E7.5. Images of E7.5 embryos shown in (j); statistics of retarded embryos shown in (k). (a,d,g,h,i,j,k) Means ± s.d. are from three independent experiments. (b,c,f,h,j) Results are representative of three independent experiments. (a,d,g,k) *: P < 0.05; **: P < 0.01; ***: P < 0.001; ns, not significant, two-tailed student’s t-test.
Supplementary information
Supplementary Information
Supplementary Protocol
Supplementary Table 1
List of sequences of RfxCas13d–BSJ-gRNA library for 762 circRNAs in this study. a, The expression (FPBcirc) of 762 circRNAs in HT29, 293FT and HeLa cells. b, The sequences of RfxCas13d/BSJ-gRNA library for 762 circRNAs.
Supplementary Table 2
CDCscreen results of negatively selected circRNAs in HT29, 293FT and HeLa cell lines. a, The CDCscreen scores of ranked circRNAs in HT29 cells (D30/D1). b, The CDCscreen scores of ranked circRNAs in 293FT cells (D30/D1). c, The CDCscreen scores of ranked circRNAs in HeLa cells (D30/D1).
Supplementary Table 3
List of CDCscreen scores of negatively selected circRNAs identified by RfxCas13d–BSJ-gRNA screening in HT29, 293FT and HeLa cell lines. a, CDCscreen scores of negatively selected circRNA in HT29 cells (D30/D1). b, CDCscreen scores of negatively selected circRNA in 293FT cells (D30/D1). c, CDCscreen scores of negatively selected circRNA in HeLa cells (D30/D1).
Supplementary Table 4
List of CDCscreen scores and FDR of negatively selected circRNAs in in vitro and in vivo screens. a, CDCscreen scores and FDR of negatively selected circRNAs in HT29 cells (in vitro). b, CDCscreen scores and FDR of negatively selected circRNAs in 293FT cells (in vitro). c, CDCscreen scores and FDR of negatively selected circRNAs in HeLa cells (in vitro). d, CDCscreen scores and FDR of negatively selected circRNAs in HT29 cells (in vivo).
Supplementary Table 5
List of validated circRNA candidates by RfxCas13d–BSJ-gRNA in HT29, HeLa and 293FT cells. The parental genes, genomic locations, FPBcirc, CDCscreen scores are listed. Most of examined candidate circRNAs can be validated by RfxCas13d with at least one gRNA in HT29, HeLa and 293FT cells, respectively.
Supplementary Table 6
List of sequences of RfxCas13d–BSJ-gRNA library for 2,908 circRNAs in this study. a, Sequences of RfxCas13d–BSJ-gRNA library for 2,908 circRNAs. b, CDCscreen scores of ranked circRNAs in HT29 in vivo (D30/D1). c, CDCscreen scores of negatively selected circRNAs in HT29 in vivo (D30/D1). d, Overlap of in vitro and in vivo CDCscreen scores of negatively selected circRNAs in HT29 (D30/D1).
Supplementary Table 7
Expression of linear genes upon circFAM120A knockdown by RfxCas13d or shRNA in 293FT cells. a, List of linear genes after circFAM120A knockdown in RfxCas13d NT and gRNA1. b, List of linear genes after circFAM120A knockdown in shRNA NT and shRNA1.
Supplementary Table 8
Expression of 24 circRNA candidates in mouse preimplantation development.
Supplementary Table 9
Screening of circRNAs with potential regulatory roles in mouse embryo preimplantation by RfxCas13d–BSJ-gRNA knockdown for each 24 circRNA candidate in mouse zygotes.
Supplementary Table 10
List of gRNA, shRNA and primer sequences used in this study. a, gRNA sequences. b, shRNA sequences. c, Primers for qRT–PCR and NB probes.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 9
Statistical source data
Source Data Extended Data Fig. 10
Statistical source data
Source Data Fig. 3
Unprocessed blots
Source Data Extended Data Fig. 1
Unprocessed blots
Source Data Extended Data Fig. 3
Unprocessed blots
Source Data Extended Data Fig. 9
Unprocessed blots and gel
Rights and permissions
About this article
Cite this article
Li, S., Li, X., Xue, W. et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat Methods 18, 51–59 (2021). https://doi.org/10.1038/s41592-020-01011-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-01011-4
This article is cited by
-
Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells
Nature Communications (2024)
-
dCas13-mediated translational repression for accurate gene silencing in mammalian cells
Nature Communications (2024)
-
Mechanisms of immune checkpoint inhibitors: insights into the regulation of circular RNAS involved in cancer hallmarks
Cell Death & Disease (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
The CRISPR-Cas13a Gemini System for noncontiguous target RNA activation
Nature Communications (2024)