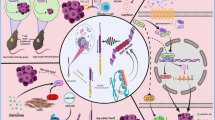
Abstract
The MYC proto-oncogenes encode a family of transcription factors that are among the most commonly activated oncoproteins in human neoplasias. Indeed, MYC aberrations or upregulation of MYC-related pathways by alternate mechanisms occur in the vast majority of cancers. MYC proteins are master regulators of cellular programmes. Thus, cancers with MYC activation elicit many of the hallmarks of cancer required for autonomous neoplastic growth. In preclinical models, MYC inactivation can result in sustained tumour regression, a phenomenon that has been attributed to oncogene addiction. Many therapeutic agents that directly target MYC are under development; however, to date, their clinical efficacy remains to be demonstrated. In the past few years, studies have demonstrated that MYC signalling can enable tumour cells to dysregulate their microenvironment and evade the host immune response. Herein, we discuss how MYC pathways not only dictate cancer cell pathophysiology but also suppress the host immune response against that cancer. We also propose that therapies targeting the MYC pathway will be key to reversing cancerous growth and restoring antitumour immune responses in patients with MYC-driven cancers.
Key points
-
The MYC oncogene is activated in the vast majority of cancers by genetic, epigenetic or post-translational mechanisms.
-
In preclinical models, inactivation of MYC can result in sustained tumour regression owing to oncogene addiction.
-
MYC activation drives cancer progression through mechanisms involving either the cell-intrinsic acquisition of hallmarks of cancer or dysregulation of the tumour microenvironment and host immune responses.
-
MYC leads to cancer initiation and maintenance by regulating the host immune system through mechanisms involving immune checkpoints, specific receptors and secreted cytokines.
-
Currently, no direct inhibitors of MYC are approved; however, many therapeutic agents targeting MYC are under development.
-
We propose that therapies targeting the MYC pathway will be key to reversing cancerous growth and restoring antitumour immune responses in patients with MYC-driven cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Schaub, F. X. et al. Pan-cancer alterations of the MYC oncogene and its proximal network across The Cancer Genome Atlas. Cell Syst. 6, 282–300.e2 (2018).
Kalkat, M. et al. MYC deregulation in primary human cancers. Genes 8, 151 (2017).
Kress, T. R., Sabò, A. & Amati, B. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 15, 593–607 (2015).
Conacci-Sorrell, M., McFerrin, L. & Eisenman, R. N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014).
Williamson, D. et al. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J. Clin. Oncol. 23, 880–888 (2005).
Williams, R. D. et al. Molecular profiling reveals frequent gain of MYCN and anaplasia-specific loss of 4q and 14q in Wilms tumor. Genes Chromosomes Cancer 50, 982–995 (2011).
Berger, A. et al. N-Myc–mediated epigenetic reprogramming drives lineage plasticity in advanced prostate cancer. J. Clin. Invest. 129, 3924–3940 (2019).
Rickman, D. S., Schulte, J. H. & Eilers, M. The expanding world of N-MYC–driven tumors. Cancer Discov. 8, 150–163 (2018).
Cheng, J. et al. Merkel cell polyomavirus recruits MYCL to the EP400 complex to promote oncogenesis. PLoS Pathog. 13, e1006668 (2017).
Ohshima, K. et al. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci. Rep. 7, 641 (2017).
Boutros, P. C. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 47, 736–745 (2015).
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
Dalla-Favera, R. et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl Acad. Sci. USA 79, 7824–7827 (1982).
Gurel, B. et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 21, 1156–1167 (2008).
Stock, C., Kager, L., Fink, F. M., Gadner, H. & Ambros, P. F. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer 28, 329–336 (2000).
Jain, M. et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297, 102–104 (2002).
Shroff, E. H. et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc. Natl Acad. Sci. USA 112, 6539–6544 (2015).
Felsher, D. W. & Bishop, J. M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4, 199–207 (1999).
Wu, C.-H. et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl Acad. Sci. USA 104, 13028–13033 (2007).
Lourenco, C. et al. Modelling the MYC-driven normal-to-tumour switch in breast cancer. Dis. Model. Mech. 12, dmm038083 (2019).
Sodir, N. M. et al. MYC instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 10, 588–607 (2020).
Soucek, L. et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 27, 504–513 (2013).
Jung, L. A. et al. OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene 36, 1911–1924 (2017).
Demma, M. J. et al. Omomyc reveals new mechanisms to inhibit the MYC oncogene.Mol. Cell. Biol. 39, e00248-19 (2019).
Weinstein, I. B. Cancer: addiction to oncogenes — the Achilles heal of cancer. Science 297, 63–64 (2002).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Carroll, P. A., Freie, B. W., Mathsyaraja, H. & Eisenman, R. N. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018).
Dang, C. V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19, 1–11 (1999).
Casey, S. C., Baylot, V. & Felsher, D. W. The MYC oncogene is a global regulator of the immune response. Blood 131, 2007–2015 (2018).
Casey, S. C., Baylot, V. & Felsher, D. W. MYC: master regulator of immune privilege. Trends Immunol. 38, 298–305 (2017).
Casacuberta-Serra, S. & Soucek, L. Myc and Ras, the Bonnie and Clyde of immune evasion. Transl. Cancer Res. 7 (Suppl. 4), S457–S459 (2018).
Baluapuri, A., Wolf, E. & Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21, 255–267 (2020).
Stine, Z. E., Walton, Z. E., Altman, B. J., Hsieh, A. L. & Dang, C. V. MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 (2015).
Bernard, S. & Eilers, M. Control of cell proliferation and growth by Myc proteins. Results Probl. Cell. Differ. 42, 329–342 (2006).
Bretones, G., Delgado, M. D. & León, J. Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516 (2015).
Levens, D. ‘You don’t muck with MYC’. Genes Cancer 1, 547–554 (2010).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017).
Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019).
Boxer, L. M. & Dang, C. V. Translocations involving c-myc and c-myc function. Oncogene 20, 5595–5610 (2001).
Drach, J. et al. The biology of multiple myeloma. J. Cancer Res. Clin. Oncol. 126, 441–447 (2000).
Dudley, J. P., Mertz, J. A., Rajan, L., Lozano, M. & Broussard, D. R. What retroviruses teach us about the involvement of c-Myc in leukemias and lymphomas. Leukemia 16, 1086–1098 (2002).
Weng, A. P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006).
He, T. C. et al. Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 (1998).
Yagi, K. et al. c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277, 854–861 (2002).
Allen-Petersen, B. L. & Sears, R. C. Mission possible: advances in MYC therapeutic targeting in cancer. BioDrugs 33, 539–553 (2019).
Sears, R. et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 (2000).
Arnold, H. K. & Sears, R. C. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol. Cell. Biol. 26, 2832–2844 (2006).
Farrell, A. S. et al. MYC regulates ductal-neuroendocrine lineage plasticity in pancreatic ductal adenocarcinoma associated with poor outcome and chemoresistance. Nat. Commun. 8, 1728 (2017).
Wang, X. et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 71, 925–936 (2011).
Reavie, L. et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 23, 362–375 (2013).
Ruvolo, P. P. The broken ‘Off’ switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 6, 87–99 (2016).
Kauko, O. et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci. Transl. Med. 10, eaaq1093 (2018).
Zhou, X. Z. & Lu, K. P. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat. Rev. Cancer 16, 463–478 (2016).
Cheng, C.-W. & Tse, E. PIN1 in cell cycle control and cancer. Front. Pharmacol. 9, 1367 (2018).
King, B. et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013).
Yeh, C.-H., Bellon, M. & Nicot, C. FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer 17, 115 (2018).
Cao, J., Ge, M.-H. & Ling, Z.-Q. Fbxw7 tumor suppressor: a vital regulator contributes to human tumorigenesis. Medicine 95, e2496 (2016).
Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699 (2000).
Edelmann, J. et al. Frequent evolution of copy number alterations in CLL following first-line treatment with FC(R) is enriched with TP53 alterations: results from the CLL8 trial. Leukemia 31, 734–738 (2017).
Yang, G. & Hurlin, P. J. MNT and emerging concepts of MNT-MYC antagonism. Genes 8, 83 (2017).
Fredlund, E., Ringnér, M., Maris, J. M. & Påhlman, S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc. Natl Acad. Sci. USA 105, 14094–14099 (2008).
Diolaiti, D., McFerrin, L., Carroll, P. A. & Eisenman, R. N. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta 1849, 484–500 (2015).
Gabay, M., Li, Y. & Felsher, D. W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 4, a014241 (2014).
Bailey, S. T. et al. MYC activation cooperates with Vhl and Ink4a/Arf loss to induce clear cell renal cell carcinoma. Nat. Commun. 8, 15770 (2017).
Vaux, D. L., Cory, S. & Adams, J. M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335, 440–442 (1988).
Teitz, T. et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat. Med. 6, 529–535 (2000).
Wu, K. J. et al. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21, 220–224 (1999).
Hofmann, J. W. et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell 160, 477–488 (2015).
Seton-Rogers, S. Driving immune evasion. Nat. Rev. Cancer 18, 67–67 (2018).
Chappell, J. & Dalton, S. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb. Perspect. Med. 3, a014381 (2013).
Pelengaris, S. & Khan, M. The many faces of c-MYC. Arch. Biochem. Biophys. 416, 129–136 (2003).
de la Cova, C., Abril, M., Bellosta, P., Gallant, P. & Johnston, L. A. Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116 (2004).
Bettess, M. D. et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 25, 7868–7878 (2005).
Evan, G. I. et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128 (1992).
Shi, Y. et al. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science 257, 212–214 (1992).
Kaczmarek, L., Hyland, J. K., Watt, R., Rosenberg, M. & Baserga, R. Microinjected c-myc as a competence factor. Science 228, 1313–1315 (1985).
Zindy, F. et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12, 2424–2433 (1998).
Karn, J., Watson, J. V., Lowe, A. D., Green, S. M. & Vedeckis, W. Regulation of cell cycle duration by c-myc levels. Oncogene 4, 773–787 (1989).
Rosenwald, I. B. Upregulated expression of the genes encoding translation initiation factors eIF-4E and eIF-2α in transformed cells. Cancer Lett. 102, 113–123 (1996).
Boon, K. et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 20, 1383–1393 (2001).
Schmidt, E. V. The role of c-myc in cellular growth control. Oncogene 18, 2988–2996 (1999).
Grewal, S. S., Li, L., Orian, A., Eisenman, R. N. & Edgar, B. A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7, 295–302 (2005).
Dong, Y., Tu, R., Liu, H. & Qing, G. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 5, 124 (2020).
Kim, J.-W. et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 24, 5923–5936 (2004).
Osthus, R. C. et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 275, 21797–21800 (2000).
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA 105, 18782–18787 (2008).
Dominguez-Sola, D. et al. Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451 (2007).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
García-Gutiérrez, L., Delgado, M. D. & León, J. MYC oncogene contributions to release of cell cycle brakes. Genes 10, 244 (2019).
Li, Z. et al. c-Myc suppression of DNA double-strand break repair. Neoplasia 14, 1190–1202 (2012).
Felsher, D. W. & Bishop, J. M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl Acad. Sci. USA 96, 3940–3944 (1999).
Kuzyk, A. & Mai, S. c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 4, a014373 (2014).
Karlsson, A. et al. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc. Natl Acad. Sci. USA 100, 9974–9979 (2003).
Vafa, O. et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9, 1031–1044 (2002).
Dang, C. V., Li, F. & Lee, L. A. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability? Cell Cycle 4, 1465–1466 (2005).
Reimann, M. et al. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood 110, 2996–3004 (2007).
Beer, S. et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2, e332 (2004).
Ray, S. et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 66, 6598–6605 (2006).
Baudino, T. A. et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 16, 2530–2543 (2002).
Shchors, K. et al. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1β. Genes Dev. 20, 2527–2538 (2006).
Dews, M. et al. The Myc–miR-17-92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246 (2010).
Giuriato, S. et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc. Natl Acad. Sci. USA 103, 16266–16271 (2006).
Rakhra, K. et al. CD4+ T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18, 485–498 (2010).
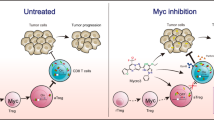
Kortlever, R. M. et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171, 1301–1315.e14 (2017).
Casey, S. C. et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016).
Swaminathan, S. et al. MYC functions as a switch for natural killer cell-mediated immune surveillance of lymphoid malignancies. Nat. Commun. 11, 2860 (2020).
Maeda, T. et al. MUC1-C induces PD-L1 and immune evasion in triple-negative breast cancer. Cancer Res. 78, 205–215 (2018).
Muthalagu, N. et al. Repression of the type I interferon pathway underlies MYC & KRAS-dependent evasion of NK & B cells in pancreatic ductal adenocarcinoma. Cancer Discov. 10, 872–887 (2020).
Dhanasekaran, R. et al. MYC and Twist1 cooperate to drive metastasis by eliciting crosstalk between cancer and innate immunity. eLife 9, e50731 (2020).
Bernards, R., Dessain, S. K. & Weinberg, R. A. N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell 47, 667–674 (1986).
Braun, J., Felsher, D. W. & Goodglick, L. A. c-myc, MHCI, and NK resistance in immunodeficiency lymphomas. Ann. N. Y. Acad. Sci. 651, 467–469 (1992).
Versteeg, R., Noordermeer, I. A., Krüse-Wolters, M., Ruiter, D. J. & Schrier, P. I. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 7, 1023–1029 (1988).
van Riggelen, J. et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 24, 1281–1294 (2010).
Reimann, M. et al. Tumor stroma-derived TGF-beta limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17, 262–272 (2010).
Xu, Y. et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 25, 301–311 (2019).
Vartuli, R. L. et al. Eya3 promotes breast tumor-associated immune suppression via threonine phosphatase-mediated PD-L1 upregulation. J. Clin. Invest. 128, 2535–2550 (2018).
Dhanasekaran, R. et al. MYC and Twist1 cooperate to drive metastasis by eliciting crosstalk between cancer and innate immunity. eLife 9, e50731 (2020).
Shachaf, C. M. et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431, 1112–1117 (2004).
Tran, P. T. et al. Combined inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS One 3, e2125 (2008).
Tran, P. T. et al. Survival and death signals can predict tumor response to therapy after oncogene inactivation. Sci. Transl. Med. 3, 103ra99 (2011).
Bui, T. V. & Mendell, J. T. Myc: maestro of microRNAs. Genes Cancer 1, 568–575 (2010).
Li, Y., Choi, P. S., Casey, S. C., Dill, D. L. & Felsher, D. W. MYC through miR-17-92 Suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 26, 262–272 (2014).
Chang, T.-C. et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40, 43–50 (2008).
Adams, C. M., Hiebert, S. W. & Eischen, C. M. Myc induces miRNA-mediated apoptosis in response to HDAC inhibition in hematologic malignancies. Cancer Res. 76, 736–748 (2016).
Zhou, L. et al. Silencing of thrombospondin-1 is critical for myc-induced metastatic phenotypes in medulloblastoma. Cancer Res. 70, 8199–8210 (2010).
Swaminathan, S. et al. MYC functions as a master switch for natural killer cell-mediated immune surveillance of lymphoid malignancies. Blood 132, 2619–2619 (2018).
Casey, S. C., Li, Y., Fan, A. C. & Felsher, D. W. Oncogene withdrawal engages the immune system to induce sustained cancer regression. J. Immunother. Cancer 2, 24 (2014).
Sharma, S. V., Fischbach, M. A., Haber, D. A. & Settleman, J. ‘Oncogenic shock’: explaining oncogene addiction through differential signal attenuation. Clin. Cancer Res. 12, 4392s–4395s (2006).
Fisher, G. H. et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15, 3249–3262 (2001).
Kohl, N. E. et al. Protein farnesyltransferase inhibitors block the growth of ras-dependent tumors in nude mice. Proc. Natl Acad. Sci. USA 91, 9141–9145 (1994).
Moody, S. E. et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, 451–461 (2002).
Hori, S. S. et al. A mathematical model of tumor regression and recurrence after therapeutic oncogene inactivation. Sci. Rep. 11, 1341 (2021).
Soucek, L. et al. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat. Med. 13, 1211–1218 (2007).
Mahauad-Fernandez, W. D. & Felsher, D. W. The Myc and Ras partnership in cancer: indistinguishable alliance or contextual relationship? Cancer Res. 80, 3799–3802 (2020).
Sun, L. et al. Gastric cancer mesenchymal stem cells derived IL-8 induces PD-L1 expression in gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death Dis. 9, 928 (2018).
Kharma, B. et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 74, 6519–6530 (2014).
Topper, M. J. et al. Epigenetic therapy Ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300.e21 (2017).
Coelho, M. A. et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 47, 1083–1099.e6 (2017).
Melaiu, O. et al. PD-L1 is a therapeutic target of the bromodomain inhibitor JQ1 and, combined with HLA Class I, a promising prognostic biomarker in neuroblastoma. Clin. Cancer Res. 23, 4462–4472 (2017).
Atsaves, V. et al. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia 31, 1633–1637 (2017).
Pai, S. et al. CD47-SIRPα signaling induces epithelial-mesenchymal transition and cancer stemness and links to a poor prognosis in patients with oral squamous cell carcinoma. Cells 8, 1658 (2019).
Kaur, S. et al. Thrombospondin-1 signaling through CD47 inhibits self-renewal by regulating c-Myc and other stem cell transcription factors. Sci. Rep. 3, 1673 (2013).
Li, W. et al. Targeting MYC activity in double-hit lymphoma with MYC and BCL2 and/or BCL6 rearrangements with epigenetic bromodomain inhibitors. J. Hematol. Oncol. 12, 73 (2019).
Kamijo, H. et al. Thrombospondin-1 promotes tumor progression in cutaneous T-cell lymphoma via CD47. Leukemia 34, 845–856 (2020).
Felsher, D. W., Rhim, S. H. & Braun, J. A murine model for B-cell lymphomagenesis in immunocompromised hosts: natural killer cells are an important component of host resistance to premalignant B-cell lines. Cancer Res. 50, 7050–7056 (1990).
El-Jawhari, J. J. et al. Blocking oncogenic RAS enhances tumour cell surface MHC class I expression but does not alter susceptibility to cytotoxic lymphocytes. Mol. Immunol. 58, 160–168 (2014).
Ji, H. et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene 25, 2105–2112 (2006).
Van Dang, C. & Kim, J.-W. Convergence of cancer metabolism and immunity: an overview. Biomol. Ther. 26, 4–9 (2018).
Gouw A. M., Hsieh A. L., Stine Z. E. & Dang C.V. In: Tumor Cell Metabolism (Mazurek S., Shoshan M. eds) 101–122 (Springer, 2015).
Dang, C. V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 3, a014217 (2013).
Gouw, A. M. et al. The MYC oncogene cooperates with sterol-regulated element-binding protein to regulate lipogenesis essential for neoplastic growth. Cell Metab. 30, 556–572.e5 (2019).
Ricciardi, S. et al. The translational machinery of human CD4 T cells is poised for activation and controls the switch from quiescence to metabolic remodeling. Cell Metab. 28, 895–906.e5 (2018).
Friesen, L. R., Gu, B. & Kim, C. H. A ligand-independent fast function of RARα promotes exit from metabolic quiescence upon T cell activation and controls T cell differentiation. Mucosal Immunol. 14, 100–112 (2021).
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Kapur, S. & Pal, A. Immune cell activation: stimulation, costimulation, and regulation of cellular activation. Immune Response Activation Immunomodul. https://doi.org/10.5772/intechopen.81568 (2019).
Shim, H. et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl Acad. Sci. USA 94, 6658–6663 (1997).
Briggs, K. J. et al. Paracrine Induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell 166, 126–139 (2016).
Bott, A. J. et al. Oncogenic Myc induces expression of glutamine synthetase through promoter demethylation. Cell Metab. 22, 1068–1077 (2015).
Gerriets, V. A. & Rathmell, J. C. Metabolic pathways in T cell fate and function. Trends Immunol. 33, 168–173 (2012).
Verbist, K. C. et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393 (2016).
Le, A. et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121 (2012).
Eberlin, L. S. et al. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc. Natl Acad. Sci. USA 111, 10450–10455 (2014).
Allison, K. E., Coomber, B. L. & Bridle, B. W. Metabolic reprogramming in the tumour microenvironment: a hallmark shared by cancer cells and T lymphocytes. Immunology 152, 175–184 (2017).
Kouidhi, S., Ben Ayed, F. & Elgaaied, A. B. Targeting tumor metabolism: a new challenge to improve immunotherapy. Front. Immunol. 9, 353 (2018).
Massó-Vallés, D. & Soucek, L. Blocking myc to treat cancer: reflecting on two decades of omomyc. Cells 9, 883 (2020).
Wolf, E. & Eilers, M. Targeting MYC proteins for tumor therapy. Annu. Rev. Cancer Biol. 4, 61–75 (2020).
McKeown, M. R. & Bradner, J. E. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 4, a014266 (2014).
Fletcher, S. & Prochownik, E. V. Small-molecule inhibitors of the Myc oncoprotein. Biochim. Biophys. Acta 1849, 525–543 (2015).
Whitfield, J. R., Beaulieu, M.-E. & Soucek, L. Strategies to inhibit myc and their clinical applicability. Front. Cell Dev. Biol. 5, 10 (2017).
Carabet, L. A., Rennie, P. S. & Cherkasov, A. Therapeutic inhibition of myc in cancer. structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci. 20, 120 (2018).
Madden, S. K., de Araujo, A. D., Gerhardt, M., Fairlie, D. P. & Mason, J. M. Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 20, 3 (2021).
Thng, D. K. H., Toh, T. B. & Chow, E. K.-H. Capitalizing on synthetic lethality of MYC to treat cancer in the digital age. Trends Pharmacol. Sci. 42, 166–182 (2021).
Cascon, A. & Robledo, M. MAX and MYC: a heritable breakup. Cancer Res. 72, 3119–3124 (2012).
Han, H. et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36, 483–497.e15 (2019).
Struntz, N. B. et al. Stabilization of the max homodimer with a small molecule attenuates Myc-driven transcription. Cell Chem. Biol. 26, 711–723.e14 (2019).
Lustig, L. C. et al. Inhibiting MYC binding to the E-box DNA motif by ME47 decreases tumour xenograft growth. Oncogene 36, 6830–6837 (2017).
Boike, L. et al. Discovery of a functional covalent ligand targeting an iIntrinsically disordered cysteine within MYC. Cell Chem. Biol. 28, 4–13.e17 (2021).
Pourdehnad, M. et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl Acad. Sci. USA 110, 11988–11993 (2013).
Wiegering, A. et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 5, 768–781 (2015).
Gustafson, W. C. et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 (2014).
Yang, D. et al. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc. Natl Acad. Sci. USA 107, 13836–13841 (2010).
Xiao, D. et al. Polo-like kinase-1 regulates myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol. Cell 64, 493–506 (2016).
Yeh, E. et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308–318 (2004).
Farrell, A. S. et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell. Biol. 33, 2930–2949 (2013).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Koh, C. M., Sabò, A. & Guccione, E. Targeting MYC in cancer therapy: RNA processing offers new opportunities. Bioessays 38, 266–275 (2016).
Ricker, J. L., Mata, J. E., Iversen, P. L. & Gattone, V. H. c-myc antisense oligonucleotide treatment ameliorates murine ARPKD. Kidney Int. 61 (Suppl. 1), S125–S131 (2002).
Yu, B. W., Nguyen, D., Anderson, S. & Allegra, C. A. Phosphorothioated antisense c-myc oligonucleotide inhibits the growth of human colon carcinoma cells. Anticancer. Res. 17, 4407–4413 (1997).
Cotter, F. E. Antisense oligonucleotides: considerations for lymphoma therapy. Hematology 1, 53–57 (1996).
Dhanasekaran, R. et al. MYC ASO impedes tumorigenesis and elicits oncogene addiction in autochthonous transgenic mouse models of HCC and RCC. Mol. Ther. Nucleic Acids 21, 850–859 (2020).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 113, 7124–7129 (2016).
Felsenstein, K. M. et al. Small molecule microarrays enable the identification of a selective, quadruplex-binding inhibitor of MYC expression. ACS Chem. Biol. 11, 139–148 (2016).
Brown, R. V., Danford, F. L., Gokhale, V., Hurley, L. H. & Brooks, T. A. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 286, 41018–41027 (2011).
Dutta, D. et al. Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 46, 5355–5365 (2018).
Hu, M.-H. et al. Discovery of a new four-leaf clover-like ligand as a potent c-MYC transcription inhibitor specifically targeting the promoter G-quadruplex. J. Med. Chem. 61, 2447–2459 (2018).
Panda, D., Saha, P., Das, T. & Dash, J. Target guided synthesis using DNA nano-templates for selectively assembling a G-quadruplex binding c-MYC inhibitor. Nat. Commun. 8, 16103 (2017).
Harbor, C. S., Dang, C. V. & Eisenman, R. N. Myc and the pathway to cancer (Cold Spring Harbor Laboratory Press, 2014).
Harrison, C. Synthetic lethality enables targeting of MYC. Nat. Rev. Drug Discov. 11, 602–602 (2012).
Kessler, J. D. et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353 (2012).
Wang, Y. et al. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell 5, 501–512 (2004).
Chen, L. et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J. Clin. Invest. 128, 446–462 (2018).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Kwiatkowski, N. et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014).
Krüger, K. et al. Multiple DNA damage-dependent and DNA damage-independent stress responses define the outcome of ATR/Chk1 targeting in medulloblastoma cells. Cancer Lett. 430, 34–46 (2018).
Murga, M. et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 18, 1331–1335 (2011).
Roeschert, I. et al. Combined inhibition of Aurora-A and ATR kinases results in regression of MYCN-amplified neuroblastoma. Nat. Cancer 2, 312–326 (2021).
Dammert, M. A. et al. MYC paralog-dependent apoptotic priming orchestrates a spectrum of vulnerabilities in small cell lung cancer. Nat. Commun. 10, 3485 (2019).
Kelly, G. L. et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 28, 58–70 (2014).
Kretzner, L. et al. Combining histone deacetylase inhibitor vorinostat with aurora kinase inhibitors enhances lymphoma cell killing with repression of c-Myc, hTERT, and microRNA levels. Cancer Res. 71, 3912–3920 (2011).
Pei, Y. et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell 29, 311–323 (2016).
Lernoux, M. et al. Novel HDAC inhibitor MAKV-8 and imatinib synergistically kill chronic myeloid leukemia cells via inhibition of BCR-ABL/MYC-signaling: effect on imatinib resistance and stem cells. Clin. Epigenet. 12, 69 (2020).
Zhao, X. et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia 27, 2341–2350 (2013).
O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005).
Dhanasekaran, R. et al. Anti-miR-17 therapy delays tumorigenesis in MYC-driven hepatocellular carcinoma (HCC). Oncotarget 9, 5517–5528 (2018).
Lai, I. et al. Lipid nanoparticles that deliver IL-12 messenger RNA suppress tumorigenesis in MYC oncogene-driven hepatocellular carcinoma. J. Immunother. Cancer 6, 125 (2018).
Pan, Y. et al. Synergistic inhibition of pancreatic cancer with anti-PD-L1 and c-Myc inhibitor JQ1. Oncoimmunology 8, e1581529 (2019).
Devi, G. R. et al. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin. Cancer Res. 11, 3930–3938 (2005).
Tolcher, A. W. et al. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J. Clin. Orthod. 33, 11006–11006 (2015).
Villanueva, M. T. Long path to MYC inhibition approaches clinical trials. Nat. Rev. Drug Discov. https://doi.org/10.1038/d41573-019-00055-2 (2019).
Shachaf, C. M. et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 68, 5132–5142 (2008).
Jung, M. et al. A myc activity signature predicts poor clinical outcomes in myc-associated cancers. Cancer Res. 77, 971–981 (2017).
Tran, D. et al. Functional genomics analysis reveals a myc signature associated with a poor clinical prognosis in liposarcomas. Am. J. Pathol. 185, 717–728 (2015).
Acknowledgements
R.D. receives grant support from the NIH (grant CA222676). A.D. has received support from Lymphoma Research Foundation. A.S.H. has received support from Lundbreck Foundation. A.M.G. has received support from the NIH (grant CA196585). D.W.F. receives grant support from the NIH (grants CA208735, CA253180, CA188383 and CA184384).
Author information
Authors and Affiliations
Contributions
R.D. and D.W.F. researched data for this manuscript. All authors contributed to the discussion of content and preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.W.F. is a consultant for Revolution Medicines, a company developing MYC pathway therapies, co-founder of Bachus and Molecular Decisions, and has had advisory roles for American Gene Technologies, Geron, Moderna and Regulus. The other authors declare no conflicts of interest.
Additional information
Peer review information
Nature Reviews Clinical Oncology thanks M. Eilers, C.M. Eischen, E.V. Prochownik and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dhanasekaran, R., Deutzmann, A., Mahauad-Fernandez, W.D. et al. The MYC oncogene — the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol 19, 23–36 (2022). https://doi.org/10.1038/s41571-021-00549-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-021-00549-2
This article is cited by
-
Investigation of the impact of bromodomain inhibition on cytoskeleton stability and contraction
Cell Communication and Signaling (2024)
-
A distinct class of pan-cancer susceptibility genes revealed by an alternative polyadenylation transcriptome-wide association study
Nature Communications (2024)
-
Combinatorial strategies to target RAS-driven cancers
Nature Reviews Cancer (2024)
-
A druggable conformational switch in the c-MYC transactivation domain
Nature Communications (2024)
-
PAF1c links S-phase progression to immune evasion and MYC function in pancreatic carcinoma
Nature Communications (2024)