Abstract
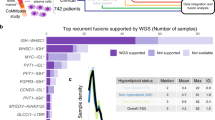
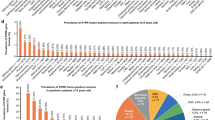
Transcript fusions as a result of chromosomal rearrangements have been a focus of attention in cancer as they provide attractive therapeutic targets. To identify novel fusion transcripts with the potential to be exploited therapeutically, we analyzed RNA sequencing, DNA copy number and gene mutation data from 4366 primary tumor samples. To avoid false positives, we implemented stringent quality criteria that included filtering of fusions detected in RNAseq data from 364 normal tissue samples. Our analysis identified 7887 high confidence fusion transcripts across 13 tumor types. Our fusion prediction was validated by evidence of a genomic rearrangement for 78 of 79 fusions in 48 glioma samples where whole-genome sequencing data were available. Cancers with higher levels of genomic instability showed a corresponding increase in fusion transcript frequency, whereas tumor samples harboring fusions contained statistically significantly fewer driver gene mutations, suggesting an important role for tumorigenesis. We identified at least one in-frame protein kinase fusion in 324 of 4366 samples (7.4%). Potentially druggable kinase fusions involving ALK, ROS, RET, NTRK and FGFR gene families were detected in bladder carcinoma (3.3%), glioblastoma (4.4%), head and neck cancer (1.0%), low-grade glioma (1.5%), lung adenocarcinoma (1.6%), lung squamous cell carcinoma (2.3%) and thyroid carcinoma (8.7%), suggesting a potential for application of kinase inhibitors across tumor types. In-frame fusion transcripts involving histone methyltransferase or histone demethylase genes were detected in 111 samples (2.5%) and may additionally be considered as therapeutic targets. In summary, we described the landscape of transcript fusions detected across a large number of tumor samples and revealed fusion events with clinical relevance that have not been previously recognized. Our results support the concept of basket clinical trials where patients are matched with experimental therapies based on their genomic profile rather than the tissue where the tumor originated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Mitelman F, Johansson B, Mertens F . The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007; 7: 233–245.
Nowell PC, Hungerford DA . A minute chromosome in human chronic granulocytic leukemia. Science 1960; 132: 1488–1501.
Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 2010; 362: 2260–2270.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566.
Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013; 368: 2385–2394.
Meyerson M, Gabriel S, Getz G . Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 2010; 11: 685–696.
Watson IR, Takahashi K, Futreal PA, Chin L . Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013; 14: 703–718.
Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337: 1231–1235.
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 2013; 45: 1459–1463.
Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3: 636–647.
Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 2014; 506: 451–455.
Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014; 343: 1010–1014.
Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G . Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics 2013; 14: 818.
Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014; 4: 216–231.
Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012; 22: 2109–2119.
Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol 2012; 227: 53–61.
Torres-Garcia W, Zheng S, Sivachenko A, Vegesna R, Wang Q, Yao R et al. PRADA: Pipeline for RNA sequencing Data Analysis. Bioinformatics 2014; 30: 2224–2226.
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009; 6: 677–681.
Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013; 45: 1134–1140.
Kalyana-Sundaram S, Shankar S, Deroo S, Iyer MK, Palanisamy N, Chinnaiyan AM et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia 2012; 14: 702–708.
Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011; 12: R6.
Kim P, Yoon S, Kim N, Lee S, Ko M, Lee H et al. ChimerDB 2.0—a knowledgebase for fusion genes updated. Nucleic Acids Res 2010; 38: D81–D85.
Mitelman F, Johansson B, Mertens F . Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet 2004; 36: 331–334.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644–648.
The Cancer Genome Atlas Research N. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med 2013; 368: 2059–2074.
Zheng S, Fu J, Vegesna R, Mao Y, Heathcock LE, Torres-Garcia W et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev 2013; 27: 1462–1472.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Shaw AT, Hsu PP, Awad MM, Engelman JA . Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013; 13: 772–787.
Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012; 367: 1694–1703.
Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19: 1469–1472.
Wells SA Jr, Gosnell JE, Gagel RF, Moley J, Pfister D, Sosa JA et al. Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J Clin Oncol 2010; 28: 767–772.
Lam ET, Ringel MD, Kloos RT, Prior TW, Knopp MV, Liang J et al. Phase II clinical trial of sorafenib in metastatic medullary thyroid cancer. J Clin Oncol 2010; 28: 2323–2330.
Hallberg B, Palmer RH . Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013; 13: 685–700.
Hojfeldt JW, Agger K, Helin K . Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov 2013; 12: 917–930.
Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M . Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012; 11: 384–400.
Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006; 106: 1794–1803.
Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, Parker S et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol 2007; 25: 3109–3115.
Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65.
Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res 2013; 73: 4711–4721.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218.
Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, Sun Z et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res 2011; 39: e100.
Gough SM, Lee F, Yang F, Walker RL, Zhu YJ, Pineda M et al. NUP98-PHF23 is a chromatin modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD domain histone reader function. Cancer Discov 2014; 4: 564–577.
Cadot B, Brunetti M, Coppari S, Fedeli S, de Rinaldis E, Dello Russo C et al. Loss of histone deacetylase 4 causes segregation defects during mitosis of p53-deficient human tumor cells. Cancer Res 2009; 69: 6074–6082.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499: 43–49.
Zhao M, Sun J, Zhao Z . TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res 2013; 41: D970–D976.
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R et al. A census of human cancer genes. Nat Rev Cancer 2004; 4: 177–183.
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G . GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12: R41.
Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013; 10: 1046–1047.
R Core Team R: A language and environment for statistical computing 2013.
Acknowledgements
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at ‘http://cancergenome.nih.gov’. This work was supported in part by the Mary K Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell), US National Cancer Institute (MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Yoshihara, K., Wang, Q., Torres-Garcia, W. et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34, 4845–4854 (2015). https://doi.org/10.1038/onc.2014.406
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.406
This article is cited by
-
The prognostic and predictive value of ESR1 fusion gene transcripts in primary breast cancer
BMC Cancer (2022)
-
Rapid and highly sensitive approach for multiplexed somatic fusion detection
Modern Pathology (2022)
-
Identification of fusions with potential clinical significance in melanoma
Modern Pathology (2022)
-
Genomic and clinical findings in myeloid neoplasms with PDGFRB rearrangement
Annals of Hematology (2022)
-
Susceptibility of cytoskeletal-associated proteins for tumor progression
Cellular and Molecular Life Sciences (2022)