Abstract
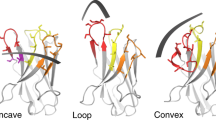
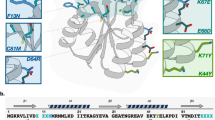
RNA crystallization and phasing represent major bottlenecks in RNA structure determination. Seeking to exploit antibody fragments as RNA crystallization chaperones, we have used an arginine-enriched synthetic Fab library displayed on phage to obtain Fabs against the class I ligase ribozyme. We solved the structure of a Fab–ligase complex at 3.1-Å resolution using molecular replacement with Fab coordinates, confirming the ribozyme architecture and revealing the chaperone's role in RNA recognition and crystal contacts. The epitope resides in the GAAACAC sequence that caps the P5 helix, and this sequence retains high-affinity Fab binding within the context of other structured RNAs. This portable epitope provides a new RNA crystallization chaperone system that easily can be screened in parallel to the U1A RNA-binding protein, with the advantages of a smaller loop and Fabs′ high molecular weight, large surface area and phasing power.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Eddy, S.R. Noncoding RNA genes. Curr. Opin. Genet. Dev. 9, 695–699 (1999).
Taft, R.J., Pheasant, M. & Mattick, J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299 (2007).
Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet. 5, e1000459 (2009).
Cooper, T.A., Wan, L. & Dreyfuss, G. RNA and disease. Cell 136, 777–793 (2009).
Hogg, J.R. & Collins, K. Structured non-coding RNAs and the RNP Renaissance. Curr. Opin. Chem. Biol. 12, 684–689 (2008).
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007).
Mattick, J.S. & Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 15 (Spec No 1), R17–R29 (2006).
Montange, R.K. & Batey, R.T. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37, 117–133 (2008).
Serganov, A. & Patel, D.J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8, 776–790 (2007).
Fedor, M.J. Comparative enzymology and structural biology of RNA self-cleavage. Ann. Rev. Biophys. 38, 271–299 (2009).
Hoogstraten, C.G. & Sumita, M. Structure-function relationships in RNA and RNP enzymes: recent advances. Biopolymers 87, 317–328 (2007).
Jinek, M. & Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 457, 405–412 (2009).
Reichow, S.L., Hamma, T., Ferré-D'Amaré, A.R. & Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 35, 1452–1464 (2007).
Doudna, J.A. & Cate, J.H. RNA structure: crystal clear? Curr. Opin. Struct. Biol. 7, 310–316 (1997).
Golden, B.L. & Kundrot, C.E. RNA crystallization. J. Struct. Biol. 142, 98–107 (2003).
Edwards, A.L., Garst, A.D. & Batey, R.T. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol. Biol. 535, 135–163 (2009).
Ke, A. & Doudna, J.A. Crystallization of RNA and RNA-protein complexes. Methods 34, 408–414 (2004).
Ferré-D'Amaré, A.R., Zhou, K. & Doudna, J.A. A general module for RNA crystallization. J. Mol. Biol. 279, 621–631 (1998).
Carrasco, N., Buzin, Y., Tyson, E., Halpert, E. & Huang, Z. Selenium derivatization and crystallization of DNA and RNA oligonucleotides for X-ray crystallography using multiple anomalous dispersion. Nucleic Acids Res. 32, 1638–1646 (2004).
Höbartner, C. et al. Syntheses of RNAs with up to 100 nucleotides containing site-specific 2′-methylseleno labels for use in X-ray crystallography. J. Am. Chem. Soc. 127, 12035–12045 (2005).
Keel, A.Y., Rambo, R.P., Batey, R.T. & Kieft, J.S. A general strategy to solve the phase problem in RNA crystallography. Structure 15, 761–772 (2007).
Robertson, M.P. & Scott, W.G. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr. D Biol. Crystallogr. D64, 738–744 (2008).
Ferré-D'Amaré, A.R. & Doudna, J.A. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 295, 541–556 (2000).
Iwata, S., Ostermeier, C., Ludwig, B. & Michel, H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 (2002).
Koide, S. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 19, 449–457 (2009).
Uysal, S. et al. Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. USA 106, 6644–6649 (2009).
Dutzler, R., Campbell, E.B., Cadene, M., Chait, B.T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002).
Tereshko, V. et al. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci. 17, 1175–1187 (2008).
Koide, S. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 19, 449–457 (2009).
Carter, P.J. Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357 (2006).
Birtalan, S. et al. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J. Mol. Biol. 377, 1518–1528 (2008).
Sidhu, S.S., Lowman, H.B., Cunningham, B.C. & Wells, J.A. Phage display for selection of novel binding peptides. Methods Enzymol. 328, 333–363 (2000).
Laird-Offringa, I.A. & Belasco, J.G. In vitro genetic analysis of RNA-binding proteins using phage display libraries. Methods Enzymol. 267, 149–168 (1996).
Ye, J.-D. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl. Acad. Sci. USA 105, 82–87 (2008).
Ekland, E.H., Szostak, J.W. & Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269, 364–370 (1995).
Bagby, S.C., Bergman, N.H., Shechner, D.M., Yen, C. & Bartel, D.P. A class I ligase ribozyme with reduced Mg2+ dependence: selection, sequence analysis, and identification of functional tertiary interactions. RNA 15, 2129–2146 (2009).
Fellouse, F.A., Wiesmann, C. & Sidhu, S.S. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. USA 101, 12467–12472 (2004).
Fellouse, F.A. et al. Molecular recognition by a binary code. J. Mol. Biol. 348, 1153 (2005).
Decanniere, K. et al. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 7, 361–370 (1999).
Desmyter, A. et al. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J. Biol. Chem. 277, 23645–23650 (2002).
Shechner, D.M. et al. Crystal structure of the catalytic core of an RNA polymerase ribozyme. Science 326, 1271–1275 (2009).
Eigenbrot, C., Randal, M., Presta, L., Carter, P. & Kossiakoff, A.A. X-ray structures of the antigen-binding domains from three variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. J. Mol. Biol. 229, 969–995 (1993).
Kossiakoff, A.A. & Koide, S. Understanding mechanisms governing protein-protein interactions from synthetic binding interfaces. Curr. Opin. Struct. Biol. 18, 499–506 (2008).
Jones, S. & Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Lunde, B.M., Moore, C. & Varani, G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 (2007).
Messias, A.C. & Sattler, M. Structural basis of single-stranded RNA recognition. Acc. Chem. Res. 37, 279–287 (2004).
Bergman, N.H., Lau, N.C., Lehnert, V., Westhof, E. & Bartel, D.P. The three-dimensional architecture of the class I ligase ribozyme. RNA 10, 176–184 (2004).
Saville, B.S. & Collins, R.A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61, 685–696 (1990).
Chao, J.A., Patskovsky, Y., Almo, S.C. & Singer, R.H. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 15, 103–105 (2008).
Hogg, J.R. & Collins, K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13, 868–880 (2007).
Keryer-Bibens, C., Barreau, C. & Osborne, H.B. Tethering of proteins to RNAs by bacteriophage proteins. Biol. Cell 100, 125–138 (2008).
Piekna-Przybylska, D., Liu, B. & Fournier, M.J. The U1 snRNA hairpin II as a RNA affinity tag for selecting snoRNP complexes. Methods Enzymol. 425, 317–353 (2007).
Kabat, E.A. & Wu, T.T. Attempts to locate complementarity-determining residues in the variable positions of light and heavy chains. Ann. NY Acad. Sci. 190, 382–393 (1971).
Fellouse, F.A. et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924 (2007).
Acknowledgements
We thank members of the Piccirilli, Koide and Kossiakoff laboratories for assistance with phage display and for insightful comments; D. Lilley and T. Wilson for helpful discussions regarding the VS ribozyme; and V. Torbeev, I. Dementieva, P. Rice and V. Tereshko for assistance with crystallography. This work was funded by Howard Hughes Medical Institute (J.A.P.), US National Institute of General Medical Sciences Medical Scientist National Research Service Award no. 5 T32 GM07281 (Y.K.) and US National Institutes of Health grants R01-GM72688 and U54-GM74946 (to S.K. and A.A.K.) and R01-GM61835 (to D.P.B.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (grant 085P1000817).
Author information
Authors and Affiliations
Contributions
All authors designed research; Y.K., E.M.D., D.M.S. and N.B.S. performed experiments; Y.K., E.M.D., D.M.S., S.K., A.A.K. and J.A.P. analyzed data; Y.K., E.M.D., D.M.S., N.B.S., D.P.B., S.K., A.A.K. and J.A.P wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Note and Supplementary Figures 1–6 (PDF 5448 kb)
Rights and permissions
About this article
Cite this article
Koldobskaya, Y., Duguid, E., Shechner, D. et al. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol 18, 100–106 (2011). https://doi.org/10.1038/nsmb.1945
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1945
This article is cited by
-
Structural basis for promiscuity in ligand recognition by yjdF riboswitch
Cell Discovery (2024)
-
Target-dependent RNA polymerase as universal platform for gene expression control in response to intracellular molecules
Nature Communications (2023)
-
Crystal structure of a highly conserved enteroviral 5′ cloverleaf RNA replication element
Nature Communications (2023)
-
Structural basis for substrate binding and catalysis by a self-alkylating ribozyme
Nature Chemical Biology (2022)
-
A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites
Nature Communications (2019)