Abstract
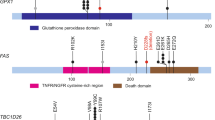
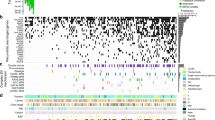
Bladder cancer is one of the most common cancers worldwide, with transitional cell carcinoma (TCC) being the predominant form. Here we report a genomic analysis of TCC by both whole-genome and whole-exome sequencing of 99 individuals with TCC. Beyond confirming recurrent mutations in genes previously identified as being mutated in TCC, we identified additional altered genes and pathways that were implicated in TCC. Notably, we discovered frequent alterations in STAG2 and ESPL1, two genes involved in the sister chromatid cohesion and segregation (SCCS) process. Furthermore, we also detected a recurrent fusion involving FGFR3 and TACC3, another component of SCCS, by transcriptome sequencing of 42 DNA-sequenced tumors. Overall, 32 of the 99 tumors (32%) harbored genetic alterations in the SCCS process. Our analysis provides evidence that genetic alterations affecting the SCCS process may be involved in bladder tumorigenesis and identifies a new therapeutic possibility for bladder cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Wu, X.R. Urothelial tumorigenesis: a tale of divergent pathways. Nat. Rev. Cancer 5, 713–725 (2005).
Gui, Y. et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011).
Cordon-Cardo, C. et al. p53 mutations in human bladder cancer: genotypic versus phenotypic patterns. Int. J. Cancer 56, 347–353 (1994).
Jebar, A.H. et al. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene 24, 5218–5225 (2005).
Cappellen, D. et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 23, 18–20 (1999).
Platt, F.M. et al. Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer. Clin. Cancer Res. 15, 6008–6017 (2009).
Cairns, P., Proctor, A.J. & Knowles, M.A. Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinoma. Oncogene 6, 2305–2309 (1991).
Hornigold, N. et al. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene 18, 2657–2661 (1999).
Solomon, D.A. et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333, 1039–1043 (2011).
Welch, J.S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Mhawech-Fauceglia, P., Cheney, R.T. & Schwaller, J. Genetic alterations in urothelial bladder carcinoma: an updated review. Cancer 106, 1205–1216 (2006).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA 104, 20007–20012 (2007).
Bertino, J.R., Goker, E., Gorlick, R., Li, W.W. & Banerjee, D. Resistance mechanisms to methotrexate in tumors. Oncologist 1, 223–226 (1996).
Williamson, M.P., Elder, P.A., Shaw, M.E., Devlin, J. & Knowles, M.A. p16 (CDKN2) is a major deletion target at 9p21 in bladder cancer. Hum. Mol. Genet. 4, 1569–1577 (1995).
Peset, I. & Vernos, I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 18, 379–388 (2008).
Singh, D. et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 (2012).
Williams, S.V., Hurst, C.D. & Knowles, M.A. Oncogenic FGFR3 gene fusions in bladder cancer. Hum. Mol. Genet. 22, 795–803 (2013).
Kiemeney, L.A. et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 42, 415–419 (2010).
Jacobs, B.L., Lee, C.T. & Montie, J.E. Bladder cancer in 2010: how far have we come? CA Cancer J. Clin. 60, 244–272 (2010).
Kops, G.J., Weaver, B.A. & Cleveland, D.W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5, 773–785 (2005).
Barber, T.D. et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 105, 3443–3448 (2008).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Koboldt, D.C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Guo, G. et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 44, 17–19 (2012).
Sjöblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006).
Getz, G. et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
Prestridge, D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 249, 923–932 (1995).
Li, L.C. & Dahiya, R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002).
Rohde, C., Zhang, Y., Reinhardt, R. & Jeltsch, A. BISMA—fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 11, 230 (2010).
Chiang, D.Y. et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods 6, 99–103 (2009).
Charchar, F.J. et al. Whole genome survey of copy number variation in the spontaneously hypertensive rat: relationship to quantitative trait loci, gene expression, and blood pressure. Hypertension 55, 1231–1238 (2010).
Yamada, N.A. et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 132, 248–254 (2011).
Jia, W. et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 14, R12 (2013).
Zhang, B., Kirov, S. & Snoddy, J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748 (2005).
Acknowledgements
This work was supported by grants from the Chinese High-Tech (863) Program (2012AA02A201 and 2012AA02A208), the Guangdong Innovative Research Team Program (2009010016), the State Key Development Program for Basic Research of China–973 Program (2011CB809203 and 2014CB745200) and the Shenzhen Municipal Government of China (JC201005260191A, CXB201108250096A, ZDSY20120615154448514 and BGI20100001).
Author information
Authors and Affiliations
Contributions
Jun Wang, Z.C., Jian Wang, H.Y., Y. Li, X. Zhang and Y.G. managed the project. X.S., S. Wu, Z. Li, A.T., X.L., Xiaokun Zhao, S.Z., M.Q., L. Xie, X. Zou, L. Xing, Z. Lv, H.M., C. Liang, J.L., C. Liu, C. Li, J.C., Y. Lai, Zheguang Lin and F. Zhou prepared the samples. P.H., P.S., F.F., Y.Y. and Xin Zhao performed the sequencing. G.G., C.C., Y.H., W.J., Q.Z., Z.Y., R.Y., Zhao Lin, S. Wan, M.L.N., M.D., S.G., Z.G., L.L., X.F., J.Y., F. Zhang, S.T. and D.T. performed the bioinformatic analysis. X.S., P.H., Z. Li, P.S., F.F., X.H., Z.J., H.C. and H.C.C. performed the validation of somatic mutations, CNAs and gene fusion events. G.G. wrote the manuscript. G.G., X.S., C.C., S. Wu, P.H. and Z. Li revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–23 and Supplementary Tables 1, 2 and 5–14 (PDF 32390 kb)
Supplementary Table 3
Predicted somatic mutations in 99 TCC patients (XLSX 857 kb)
Supplementary Table 4
A list of all confirmed somatic mutations detected in 99 TCC patients (XLSX 83 kb)
Rights and permissions
About this article
Cite this article
Guo, G., Sun, X., Chen, C. et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 45, 1459–1463 (2013). https://doi.org/10.1038/ng.2798
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2798
This article is cited by
-
The synergism of SMC1A cohesin gene silencing and bevacizumab against colorectal cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Incidental detection of FGFR3 fusion via liquid biopsy leading to earlier diagnosis of urothelial carcinoma
npj Precision Oncology (2023)
-
TRAIL-mediated signaling in bladder cancer: realization of clinical efficacy of TRAIL-based therapeutics in medical oncology
Medical Oncology (2023)
-
Novel Method for Detection of PIK3CA Mutations in Circulating Tumor DNA of Patients with Colorectal Cancer
Applied Biochemistry and Biotechnology (2023)
-
Alterations of cohesin complex genes in acute myeloid leukemia: differential co-mutations, clinical presentation and impact on outcome
Blood Cancer Journal (2023)