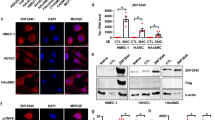
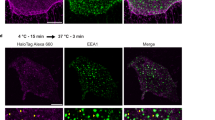
Abstract
Notch signalling is a key intercellular communication mechanism that is essential for cell specification and tissue patterning, and which coordinates critical steps of blood vessel growth1,2,3 . Although subtle alterations in Notch activity suffice to elicit profound differences in endothelial behaviour and blood vessel formation2,3 , little is known about the regulation and adaptation of endothelial Notch responses. Here we report that the NAD+-dependent deacetylase SIRT1 acts as an intrinsic negative modulator of Notch signalling in endothelial cells. We show that acetylation of the Notch1 intracellular domain (NICD) on conserved lysines controls the amplitude and duration of Notch responses by altering NICD protein turnover. SIRT1 associates with NICD and functions as a NICD deacetylase, which opposes the acetylation-induced NICD stabilization. Consequently, endothelial cells lacking SIRT1 activity are sensitized to Notch signalling, resulting in impaired growth, sprout elongation and enhanced Notch target gene expression in response to DLL4 stimulation, thereby promoting a non-sprouting, stalk-cell-like phenotype. In vivo, inactivation of Sirt1 in zebrafish and mice causes reduced vascular branching and density as a consequence of enhanced Notch signalling. Our findings identify reversible acetylation of the NICD as a molecular mechanism to adapt the dynamics of Notch signalling, and indicate that SIRT1 acts as rheostat to fine-tune endothelial Notch responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
20 April 2011
In Fig. 1, panel l was corrected; in Fig. 2, panels d, g, h and j were corrected.
References
Kopan, R. & Ilagan, M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009)
Roca, C. & Adams, R. H. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 21, 2511–2524 (2007)
Phng, L. K. & Gerhardt, H. Angiogenesis: a team effort coordinated by notch. Dev. Cell 16, 196–208 (2009)
Milne, J. C. et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 (2007)
Kurooka, H. & Honjo, T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275, 17211–17220 (2000)
Oswald, F. et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21, 7761–7774 (2001)
Kim, M. Y. et al. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol. Cell. Biol. 27, 6506–6519 (2007)
Potente, M. et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 21, 2644–2658 (2007)
Hellström, M. et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445, 776–780 (2007)
Ridgway, J. et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 444, 1083–1087 (2006)
Suchting, S. et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl Acad. Sci. USA 104, 3225–3230 (2007)
Lobov, I. B. et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc. Natl Acad. Sci. USA 104, 3219–3224 (2007)
Phng, L. K. et al. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev. Cell 16, 70–82 (2009)
Benedito, R. et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 137, 1124–1135 (2009)
Bentley, K., Mariggi, G., Gerhardt, H. & Bates, P. A. Tipping the balance: robustness of tip cell selection, migration and fusion in angiogenesis. PLoS Comput. Biol. 5, e1000549 (2009)
Siekmann, A. F. & Lawson, N. D. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 445, 781–784 (2007)
Leslie, J. D. et al. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development 134, 839–844 (2007)
Jakobsson, L. et al. Heparan sulfate in trans potentiates VEGFR-mediated angiogenesis. Dev. Cell 10, 625–634 (2006)
Jakobsson, L. et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nature Cell Biol. 12, 943–953 (2010)
Picard, F. et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429, 771–776 (2004)
Hisahara, S. et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl Acad. Sci. USA 105, 15599–15604 (2008)
Takata, T. & Ishikawa, F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301, 250–257 (2003)
Prozorovski, T. et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nature Cell Biol. 10, 385–394 (2008)
Kitamura, T. et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Invest. 117, 2477–2485 (2007)
Donmez, G., Wang, D., Cohen, D. E. & Guarente, L. SIRT1 suppresses β-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142, 320–332 (2010)
Finkel, T., Deng, C. X. & Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 (2009)
Potente, M. et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 115, 2382–2392 (2005)
North, B. J. et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 (2003)
Kim, J. E., Chen, J. & Lou, Z. DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 (2008)
Shevchenko, A. et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 1, 2856–2860 (2006)
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 (2007)
Olsen, J. V. et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 (2009)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 (2008)
Vintersten, K. et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis 40, 241–246 (2004)
Cheng, H. L. et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl Acad. Sci. USA 100, 10794–10799 (2003)
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002)
Acknowledgements
We are thankful to F. W. Alt, R. Kopan, Z. Lou, E. Seto, S. L. Berger, S. Diane Hayward, S. McMahon, G. Thurston and N. D. Lawson for reagents and to I. Dikic for comments. This work was supported by grants from the DFG (PO1306/1-1, SFB 834/A6 and Exc 147/1). F.D. was supported by the Interuniversity Attraction Poles Program–Belgian Science Policy (IUAP-BELSPO PVI/28). R.M. is supported by the Sidney Kimmel Cancer Research Foundation, a New Investigator Grant from the Massachusetts Life Sciences Center, an AFAR Research Grant and NIH grants (R01DK088190-01A1 and R01GM093072-01). H.G. is supported by Cancer Research UK, the European Molecular Biology Organisation Young Investigator Programme, and The Lister Institute of Preventive Medicine. H.G. and K.B. are supported by the Fondation Leducq Transatlantic Network of Excellence ARTEMIS. C.A.F. is supported by the Marie Curie FP7 People initiative. G.D. and M.M. thank F. Pezzimenti for fish care and technical help, and AIRC (Associazione Italiana per la Ricerca sul Cancro) for financial support.
Author information
Authors and Affiliations
Contributions
V.G. and M.P. designed and guided research. V.G., G.D., C.A.F., M.K., L.-K.P., K.B., L.T., F.D., M.H.H.S., B.Z., R.P.B., M.M., H.G. and M.P. performed research. V.G., G.D., C.A.F., M.K., L.-K.P., K.B., L.T., F.D., M.M., H.G. and M.P. analysed data. R.M., C.H.W. and T.B. provided reagents and/or technical support. T.B., A.M.Z. and S.D. gave conceptual advice. V.G., H.G. and M.P. wrote the paper. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-16 with legends. (PDF 24705 kb)
Supplementary Movie 1
This movie shows simulation of a Sirt1 wild-type vessel. Tip cell selection occurs fast, movie represents 1 hour and 52 minutes (450 model time steps). Colour indicates NICD levels, purple - low, green – high (MOV 1098 kb)
Supplementary Movie 2
This movie shows simulation of a Sirt1-deficient vessel. Tip cell selection occurs slowly after initial oscillations in NICD levels. Colour indicates NICD levels, purple - low, green - high. (MOV 2410 kb)
Rights and permissions
About this article
Cite this article
Guarani, V., Deflorian, G., Franco, C. et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 473, 234–238 (2011). https://doi.org/10.1038/nature09917
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09917
This article is cited by
-
Non-canonical non-genomic morphogen signaling in anucleate platelets: a critical determinant of prothrombotic function in circulation
Cell Communication and Signaling (2024)
-
SIRT1 mediates the inhibitory effect of Dapagliflozin on EndMT by inhibiting the acetylation of endothelium Notch1
Cardiovascular Diabetology (2023)
-
The role of mitochondria in rheumatic diseases
Nature Reviews Rheumatology (2022)
-
A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth
Nature Metabolism (2022)
-
Control of endothelial quiescence by FOXO-regulated metabolites
Nature Cell Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.