Abstract
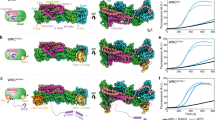
Members of the Wiskott–Aldrich syndrome protein (WASP) family control cytoskeletal dynamics by promoting actin filament nucleation with the Arp2/3 complex. The WASP relative WAVE regulates lamellipodia formation within a 400-kilodalton, hetero-pentameric WAVE regulatory complex (WRC). The WRC is inactive towards the Arp2/3 complex, but can be stimulated by the Rac GTPase, kinases and phosphatidylinositols. Here we report the 2.3-ångstrom crystal structure of the WRC and complementary mechanistic analyses. The structure shows that the activity-bearing VCA motif of WAVE is sequestered by a combination of intramolecular and intermolecular contacts within the WRC. Rac and kinases appear to destabilize a WRC element that is necessary for VCA sequestration, suggesting the way in which these signals stimulate WRC activity towards the Arp2/3 complex. The spatial proximity of the Rac binding site and the large basic surface of the WRC suggests how the GTPase and phospholipids could cooperatively recruit the complex to membranes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Takenawa, T. & Suetsugu, S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nature Rev. Mol. Cell Biol. 8, 37–48 (2007)
Pollitt, A. Y. & Insall, R. H. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J. Cell Sci. 122, 2575–2578 (2009)
Padrick, S. B. & Rosen, M. K. Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem. 79, 707–735 (2010)
Kim, A. S., Kakalis, L. T., Abdul-Manan, N., Liu, G. A. & Rosen, M. K. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 404, 151–158 (2000)
Padrick, S. B. et al. Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426–438 (2008)
Eden, S., Rohatgi, R., Podtelejnikov, A. V., Mann, M. & Kirschner, M. W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793 (2002)
Stovold, C. F., Millard, T. H. & Machesky, L. M. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 6, 11 (2005)
Vartiainen, M. K. & Machesky, L. M. The WASP-Arp2/3 pathway: genetic insights. Curr. Opin. Cell Biol. 16, 174–181 (2004)
Napoli, I. et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054 (2008)
Schenck, A. et al. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 38, 887–898 (2003)
Kobayashi, K. et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273, 291–295 (1998)
Oikawa, T. et al. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nature Cell Biol. 6, 420–426 (2004)
Ismail, A. M., Padrick, S. B., Chen, B., Umetani, J. & Rosen, M. K. The WAVE regulatory complex is inhibited. Nature Struct. Mol. Biol. 16, 561–563 (2009)
Lebensohn, A. M. & Kirschner, M. W. Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 36, 512–524 (2009)
Sossey-Alaoui, K., Li, X. & Cowell, J. K. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem. 282, 26257–26265 (2007)
Stuart, J. R., Gonzalez, F. H., Kawai, H. & Yuan, Z. M. c-Abl interacts with the WAVE2 signaling complex to induce membrane ruffling and cell spreading. J. Biol. Chem. 281, 31290–31297 (2006)
Leng, Y. et al. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl Acad. Sci. USA 102, 1098–1103 (2005)
Miyamoto, Y., Yamauchi, J. & Tanoue, A. Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase Fyn. J. Neurosci. 28, 8326–8337 (2008)
Ardern, H. et al. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil. Cytoskeleton 63, 6–13 (2006)
Dai, Z. & Pendergast, A. M. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9, 2569–2582 (1995)
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008)
Soding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, 244–248 (2005)
Jia, D. et al. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl Acad. Sci. USA 107, 10442–10447 (2010)
Derivery, E. et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 (2009)
Gomez, T. S. & Billadeau, D. D. A. FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 (2009)
Goley, E. D., Rodenbusch, S. E., Martin, A. C. & Welch, M. D. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol. Cell 16, 269–279 (2004)
Marchand, J. B., Kaiser, D. A., Pollard, T. D. & Higgs, H. N. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nature Cell Biol. 3, 76–82 (2001)
Chereau, D. et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl Acad. Sci. USA 102, 16644–16649 (2005)
Panchal, S. C., Kaiser, D. A., Torres, E., Pollard, T. D. & Rosen, M. K. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nature Struct. Biol. 10, 591–598 (2003)
Stradal, T. E. et al. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14, 303–311 (2004)
Steffen, A. et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759 (2004)
Weiner, O. D. et al. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 4, e38 (2006)
Ryu, J. R., Echarri, A., Li, R. & Pendergast, A. M. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol. Cell. Biol. 29, 1735–1748 (2009)
Nakao, S., Platek, A., Hirano, S. & Takeichi, M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J. Cell Biol. 182, 395–410 (2008)
Nowicki, S. T. et al. The Prader-Willi phenotype of fragile X syndrome. J. Dev. Behav. Pediatr. 28, 133–138 (2007)
Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008)
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006)
Dodson, E. J., Winn, M. & Ralph, A. Collaborative Computational Project number 4: providing programs for protein crystallography. Methods Enzymol. 277, 620–633 (1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Otwinowski, Z. M. W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Zhang, K. Y., Cowtan, K. & Main, P. Combining constraints for electron-density modification. Methods Enzymol. 277, 53–64 (1997)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Borek, D., Ginell, S. L., Cymborowski, M., Minor, W. & Otwinowski, Z. The many faces of radiation-induced changes. J. Synchrotron Radiat. 14, 24–33 (2007)
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, 375–383 (2007)
Nolz, J. C. et al. The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr. Biol. 16, 24–34 (2006)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, 299–302 (2005)
Acknowledgements
We thank B. Chen for providing samples of full-length WRC and VCA polypeptide for electron microscopy and some biochemical analyses, C. Pak for helping with the total internal reflection fluorescence experiment, D. Tomchick and C. Brautigam for technical assistance and N. Grishin for assistance with sequence analysis and discussion. Research was supported by fellowships from the Cancer Research Institute and the NIH (1F32-GM06917902) to Z.C. and S.B.P., respectively, an Allergic Diseases Training grant (AI07047) to T.S.G., grants from the NIH to D.D.B. (R01-AI065474), Z.O. (R01-GM053163) and M.K.R. (R01-GM056322), a grant from the Welch Foundation to M.K.R. (I-1544) and the Howard Hughes Medical Institute. D.D.B. is a Leukemia and Lymphoma Society Scholar. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US DOE under contract DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
M.K.R. oversaw the project. Z.C., A.I., S.B.P. and J.U. developed the WRC reconstitution. D.B., Z.C. and Z.O. determined the structure of the WRC. Z.C. performed the biochemical experiments. D.D.B. and T.S.G. performed the cellular experiments. Z.M. performed the electron microscopy experiments. .Z.C., Z.O. and M.K.R. analysed the WRC structure. D.B., Z.C., Z.O., S.B.P. and M.K.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-15 with legends and additional references. (PDF 17463 kb)
Rights and permissions
About this article
Cite this article
Chen, Z., Borek, D., Padrick, S. et al. Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538 (2010). https://doi.org/10.1038/nature09623
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09623
This article is cited by
-
Orchestration of synaptic functions by WAVE regulatory complex-mediated actin reorganization
Experimental & Molecular Medicine (2023)
-
Time-dependent inhibition of Rac1 in the VTA enhances long-term aversive memory: implications in active forgetting mechanisms
Scientific Reports (2023)
-
Small extracellular vesicles promote invadopodia activity in glioblastoma cells in a therapy-dependent manner
Cellular Oncology (2023)
-
Regulation of the Scar/WAVE complex in migrating cells: A summary of our understanding
Journal of Biosciences (2023)
-
Role of NCKAP1 in the Defective Phagocytic Function of Microglia-Like Cells Derived from Rapidly Progressing Sporadic ALS
Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.