Abstract
The histone methyltransferase G9a is overexpressed in a variety of cancer types, including pancreatic adenocarcinoma, and promotes tumor invasiveness and metastasis. We recently reported the discovery of BRD4770, a small-molecule inhibitor of G9a that induces senescence in PANC-1 cells. We observed that the cytotoxic effects of BRD4770 were dependent on genetic background, with cell lines lacking functional p53 being relatively resistant to compound treatment. To understand the mechanism of genetic selectivity, we used two complementary screening approaches to identify enhancers of BRD4770. The natural product and putative BH3 mimetic gossypol enhanced the cytotoxicity of BRD4770 in a synergistic manner in p53-mutant PANC-1 cells but not in immortalized non-tumorigenic pancreatic cells. The combination of gossypol and BRD4770 increased LC3-II levels and the autophagosome number in PANC-1 cells, and the compound combination appears to act in a BNIP3 (B-cell lymphoma 2 19-kDa interacting protein)-dependent manner, suggesting that these compounds act together to induce autophagy-related cell death in pancreatic cancer cells.
Similar content being viewed by others
Main
The clinical outcome of cancer treatment is highly variable, partially due to the genetic variation of cancer genomes. Mutations in genes regulating apoptosis and DNA repair pathways confer resistance to a broad range of cytotoxic drugs. For instance, loss of functional p53 protein is a predictor of failure to respond to radiotherapy and chemotherapy in various human cancers.1, 2 The pairing of conventional chemotherapeutic drugs with compounds targeting epigenetic mechanisms, usually inhibitors of histone deacetylases and DNA methyltransferases (DNMTs), has been a useful strategy to combat this resistance.3 The discovery of small-molecule probes against different types of chromatin-modifying enzymes, such as histone methyltransferases (HMTs), enables investigation into whether these enzymes constitute cancer dependencies in the context of single-agent treatment or in combinations.
Previous studies suggest a role for the HMT G9a in tumorigenesis and cancer progression, for example, by increasing chromosome instability and promoting metastasis.4, 5 G9a and G9a-like protein (GLP) are the primary HMTs responsible for histone H3 lysine 9 methylation in euchromatic DNA.6 However, G9a also methylates lysine residues on non-histone protein substrates, such as p53, inhibiting its tumor suppressive activity.7 We recently reported the discovery of BRD4770, an S-adenosylmethionine mimetic inhibitor of G9a that promotes senescence in PANC-1 cells, which lack functional p53 and p16.8 Although BRD4770 shows little toxicity in this genetic context, it is possible that its induction of senescence pathways may give rise to new vulnerabilities that could be targeted by small molecules in combination with BRD4770.
To identify small molecules that, in combination with BRD4770, can promote cell death even in the absence of p53, we performed a pilot screening of known probes and drugs that target cancer-relevant pathways using two assay readouts of cell viability in PANC-1 cells. Here, we show that gossypol, a natural product isolated from cottonseeds, sensitizes PANC-1 cells to BRD4770 and interacts in a synergistic manner to induce cell death. No cytotoxic effects were observed in hHPNE, an hTERT-immortalized but non-cancerous pancreatic duct epithelial cell line expressing wild-type p16, p53, and K-RAS.9 Gossypol induces autophagy, an evolutionarily conserved pathway for maintaining cellular homeostasis by eliminating excessive or unnecessary proteins and injured or aged organelles in normal cells.10 Autophagy has been associated with tumor formation and progression; both inducers and inhibitors of autophagy can cause cancer-cell death, including cancer cells resistant to chemotherapy-induced apoptosis.11, 12 We found that LC3-II levels and the number of autophagosomes were increased by the compound combination in PANC-1 cells. Moreover, we observed an upregulation of BNIP3 (B-cell lymphoma 2 (BCL2) 19-kDa interacting protein) expression by inhibition of G9a, a phenomenon likely to be involved in this synergistic cell death. Together, these data suggest an additional role for inhibitors of HMTs in cancer-cell death.
Results
Cancer-cell sensitivity to BRD4770 depends on p53 status
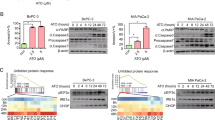
To investigate whether p53 status in cancer cell lines is responsible for differential sensitivity to BRD4770 treatment, we tested BRD4770 in five human cancer cell lines. MCF7 breast and HPAC pancreatic adenocarcinoma cells have wild-type TP53 and express functional p53 protein; PANC-1 pancreatic adenocarcinoma cells have only one allele of TP53, which contains an R273C mutation in the DNA-binding region; HeLa cervical adenocarcinoma cells have wild-type TP53 but no functional p53 protein due to rapid degradation; and PC-3 prostate adenocarcinoma cells have both TP53 alleles deleted. The cell lines without functional p53 protein were relatively more resistant to BRD4770-induced cell death, as measured by ATP levels (Figure 1a). The modified MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay13 data also suggest a lower survival rate of cell lines with functional p53 upon BRD4770 treatment (Supplementary Figure S1). Moreover, caspase-3/7 activity, indicative of apoptosis, was induced only in p53-positive cell lines (Figure 1b). To determine whether the p53 pathway was activated upon BRD4770 treatment, we examined the post-translational modifications of p53 after 3-day compound treatment. An increase in p53 acetylation and phosphorylation indicated its activation by compound treatment, although total p53 protein levels were unaffected (Figure 1c, Supplementary Figure S2A). We then analyzed the effect of BRD4770 on the expression of eight direct downstream targets of p53 by real-time PCR. Six of the eight genes were upregulated in MCF7, and four genes were upregulated in HPAC cells (both with wild-type p53), whereas none of the eight genes were increased in any of the p53-mutant cell lines (Figure 1d). Consistent with the mutational status in the DNA-binding domain of p53, BRD4770-treated PANC-1 cells were unable to induce expression of downstream p53 targets (Figure 1d). A luciferase reporter gene assay for p53 activity was performed in both MCF7 and PANC-1 cells; p53 activity was induced by BRD4770 in a dose-dependent manner only in wild-type p53 MCF7 cells (Supplementary Figures S2A and B).
Lack of functional p53 renders cancer cells more resistant to the G9a inhibitor BRD4770. (a) Cellular ATP levels after 3-day treatment of five cancer cell lines (p53+, functional protein; p53−, lack of functional protein) with BRD4770. Data represent the mean and standard error of six biological replicates. (b) Caspase-3/7 activities in five cancer cell lines were measured after 3-day treatment with BRD4770. Results were normalized by cellular ATP levels, and data represent the mean and standard error of six independent replicates. (c) Western blot analysis of p53, phospho-p53 (Ser15), and acetyl-p53 (Lys382) levels in PANC-1 cells after 3-day treatment with BRD4770. Tubulin was used as an internal loading control. (d) Gene expression analysis, qPCR, of eight transcriptional targets of p53. Data were normalized to control genes GAPDH and actin. A heatmap illustrates the fold change over DMSO controls
Identification of small-molecule enhancers of BRD4770
To identify small molecules that overcome resistance of p53-mutant cell lines to BRD4770, we performed a pilot screening in PANC-1 cells using two assay readouts. First, we tested 198 bioactive compounds in four doses for their effects on cellular ATP levels. Three of these compounds enhanced the inhibitory effects of BRD4770 on ATP levels in PANC-1 cells (Supplementary Figure S3). Second, we assessed 92 bioactive compounds for their effects on cellular metabolism using the Phenotype Microarray platform (Biolog Inc., Hayward, CA, USA).13 Four compounds enhanced cell death, as measured by metabolic dye reduction (Supplementary Figure S4). None of these hit compounds enhanced cell death in hHPNE, which expresses relatively low levels of G9a (Supplementary Figure S5).
The natural product gossypol was a common hit in both assays and showed selectivity between PANC-1 and hHPNE cells (Supplementary Figure S3). We measured cellular ATP levels after 3-day treatment with different combinations of BRD4770 and gossypol in both PANC-1 and hHPNE cells. Gossypol treatment greatly enhanced cell death in PANC-1 cells, whereas no effect was observed in hHPNE cells (Figures 2a and c). Calculation of synergy revealed the strongest effect to be the combination of 1 μM gossypol and 2.5 μM BRD4770, with a combination index (CI) of 0.23 (Figure 2b), where CI<1 indicates synergistic interaction. All combinations showed a simple additive effect in hHPNE cells (Figure 2d). We also tested UNC0638, a structurally unrelated inhibitor of G9a and GLP,14 in combination with gossypol in both PANC-1 and hHPNE cells. The interaction of UNC0638 with gossypol was very similar to that of BRD4770, with a CI of 0.43 (Supplementary Figure S6), suggesting that gossypol specifically enhances G9a inhibition in p53-mutant cells to cause cell death. However, the addition of gossypol did not induce further caspase activation in wild-type p53 MCF7 cells (Supplementary Figure S7).
Gossypol and BRD4770 synergize to induce cell death in PANC-1 cells, but not in hHPNE cells. (a) Cellular ATP levels in PANC-1 after 3-day treatment with the indicated concentrations of gossypol and BRD4770. (b) A heatmap representing the CI for each combination of gossypol and BRD4770 in PANC-1 cells. (c) Cellular ATP levels in hHPNE after 3-day treatment with the same combination of gossypol and BRD4770. Data represent the mean and standard error of four independent replicates for each combination. (d) A heatmap representing the CI of the same compound combinations in hHPNE cells
BRD4770 and gossypol increase autophagosome number in cells
We then sought to understand the mechanism of cell death induced by gossypol and BRD4770. In contrast to p53-positive cell lines, PANC-1 cells did not undergo apoptosis in response to gossypol and BRD4770, either alone or in combination (Supplementary Figure S8). As gossypol induces autophagy in several different cancer cell lines,15, 16, 17, 18, 19 we investigated whether gossypol has similar effects in PANC-1 cells. LC3-II, a protein recruited to autophagosomes, is widely used as a marker of autophagy.20 LC3-II protein levels in PANC-1 cells increased significantly in response to treatment with both BRD4770 and gossypol (Figure 3a). We then used HeLa cells stably transfected with a plasmid encoding mCherry-eGFP-LC3, which enabled us to visualize autophagosomes in the red channel. We confirmed that synergistic cell death was also induced by the combination of gossypol and BRD4770 in this cell line (Supplementary Figure S9). Accordingly, we observed an increased intensity and punctate pattern of LC3 expression with a combination of 2 μM gossypol and 2.5 μM BRD4770, whereas no strong fluorescent signal was observed in cells treated with those concentrations of gossypol or BRD4770 alone (Figure 3b). Addition of the lysosomal protease inhibitors E64d and pepstatin, which block the degradation of autophagosomes,21 further increased LC3-II levels in both PANC-1 and HeLa cells (Supplementary Figure S10), suggesting that the compound combination might increase autophagic flux rather than acting as lysosomal inhibitors.
Gossypol and BRD4770 induce LC3-II expression and autophagosome formation in tumor cell lines. (a) LC3-II and BNIP3 protein levels in PANC-1 cells treated for 3 days with the indicated concentrations of gossypol and BRD4770. Tubulin was used as an internal loading control. (b) LC3 expression in HeLa cells stably expressing mCherry-eGFP-LC3. Cells were treated for 24 h with the indicated concentrations of gossypol alone and in combination with 2.5 μM BRD4770. Scale bar=20 μm
A number of small-molecule inducers of autophagy, acting through mTOR-dependent and mTOR -independent pathways, have been identified.22 To determine whether the interaction between BRD4770 and gossypol is a general property of autophagy-related cell death, we treated PANC-1 cells with a combination of BRD4770 and well-known autophagy modulators – rapamycin, tamoxifen, clonidine, carbamazepine, verapamil, loperamide, or a PI3Kγ inhibitor. None of these compounds had a synergistic effect with BRD4770 (Supplementary Figure S11). Because gossypol is reported to be a BCL2 homology domain 3 (BH3) mimetic, we also tested two BCL2 inhibitors in combination with BRD4770: ABT-737, a BH3 mimetic,23, 24 and HA14-1.25 ABT-737 displayed a moderate synergistic effect with BRD4770 at high concentrations, whereas HA14-1 was only an additive with BRD4770 in PANC-1 cells (Supplementary Figure S12). These results suggest that BRD4770 does not generally synergize with either inducers of autophagy or inhibitors of BCL2.
BRD4770 induces BNIP3 expression
To further investigate the interactions between gossypol and BRD4770, we measured the expression of a panel of 84 genes important to autophagy. Treatment with both compounds increased eight of the transcripts greater than twofold over dimethylsulfoxide (DMSO) controls and decreased expression of two genes (Figure 4a). In particular, BNIP3 was induced 7.1-fold compared with DMSO controls and was increased 3.6-fold upon addition of BRD4770 to gossypol. BNIP3 and BNIP3L are BH3-only proapoptotic members of the BCL2 family,26 and increased levels of BNIP3 and BNIP3L are responsible for hypoxia-induced autophagy in a variety of cell types.27, 28 BRD4770 treatment alone increased protein levels of BNIP3 and BNIP3L, whereas two other proteins involved in gossypol-induced autophagy, beclin1 and BCL-xL,29 were relatively unaffected by BRD4770 treatment (Figure 4b). At higher concentrations of BRD4770 (10 μM) or UNC0638 (5 μM), we observed increased LC3 levels (Supplementary Figure S13).
BNIP3 mRNA and protein levels are increased upon compound treatment. (a) Fold change in gene expression in PANC-1 cells treated for 3 days with 2.5 μM gossypol alone or the combination of 2.5 μM gossypol and 2.5 μM BRD4770. The 10 gene transcripts that showed the greatest fold induction between the two conditions are shown. The total RNA was extracted, and the 84 key genes involved in autophagy were profiled by a 384-well format human autophagy pathway-focused expression real-time PCR array. Six housekeeping genes were used as internal controls for data normalization (see the Materials and Methods section). Data for BRD4770 treatment were generated by gene expression profiling (see the Materials and Methods section). (b) Protein levels of BNIP3, BNIP3L, beclin1, and BCL-xL were assessed in PANC-1 cells treated for 3 days with the indicated concentrations of BRD4770. BNIP3 was observed as a 30-kDa monomer and a 60-kDa dimer. Tubulin was used as an internal loading control
Overexpression of BNIP3 increases LC3-II levels and autophagosome formation in the presence of gossypol
Although increased levels of BNIP3 and BNIP3L induce autophagy-related cell death in many cancer cells,27, 28, 30 it is unclear whether expression of BNIP3 and BNIP3L is responsible for the increase of the autophagosome number in PANC-1 cells. Thus, we overexpressed human BNIP3 and BNIP3L, individually and in combination, in PANC-1 and mCherry-eGFP-LC3 HeLa cells. Transfected cells were treated with 2.5 μM gossypol for an additional 24 h. We observed an increase in LC3-II protein levels in cells overexpressing either BNIP3 alone or the combination of BNIP3 and BNIP3L (Figure 5a). We also observed increased LC3 fluorescence in mCherry-eGFP-LC3 HeLa cells when BNIP3 was overexpressed (Figure 5b). BNIP3L alone was not sufficient to induce LC3-II levels in either cell line (Figures 5a and b), suggesting that the increased expression of BNIP3 is sufficient to increase the autophagosome number in PANC-1 cells.
BNIP3 overexpression induces LC3-II expression and autophagosome formation in the presence of gossypol. (a) PANC-1 cells were transiently transfected with BNIP3 and BNIP3L, either individually or in combination. The control cells were transfected with an empty vector. After 24 h, the cells were treated with 2.5 μM gossypol for another 24 h. Tubulin served as an internal loading control. (b) Fluorescence microscopy of mCherry-eGFP-LC3 HeLa cells treated as described in panel (a). Scale bar=20 μm
Both genetic and small-molecule inhibition of the HMT G9a induce BNIP3 expression and synergistic cell death with gossypol
We then sought to determine whether reduced expression of G9a or GLP, the putative targets of BRD4770, was sufficient for the induction of autophagy. To that end, we combined gossypol treatment with genetic knockdown of G9a and GLP in PANC-1 cells (Figure 6a). Cellular ATP levels were modestly decreased by gossypol, and this effect was enhanced by knockdown of either G9a alone or both G9a and GLP (Figure 6b). Both individual and combinatorial knockdown of G9a and GLP induced the expression of LC3-II, BNIP3, and BNIP3L (Figure 6c). The effects on BNIP3L were strongest in the double knockdown.
Knockdown of G9a and GLP expression induces BNIP3 expression and cell death. PANC-1 cells were transiently transfected with siRNA against G9a and GLP, both individually and in combination. Scrambled siRNA was used as the negative control. (a) Quantification of the level of knockdown for G9a and GLP by real-time PCR. GAPDH and actin were used to normalize the data. (b) Cellular ATP levels were measured after 3-day treatment with gossypol and either 1.25 μM BRD4770 or siRNA knockdown of G9a and GLP. Data represent the mean and standard error of six independent replicates for each combination. (c) Western blot analysis to examine the protein levels of LC3, BNIP3, and BNIP3L 3 days after transfection. Tubulin served as an internal loading control
Discussion
This study reveals that p53 inactivation contributes to cancer-cell resistance to BRD4770, a previously identified inhibitor of the HMT G9a. Using small-molecule screens that assess ATP levels and metabolic activity, we found the natural product gossypol to enhance BRD4770 cytotoxicity significantly in p53-mutant cells in a manner consistent with the induction of autophagy. The interaction between gossypol and BRD4770 was synergistic, suggesting that the compounds act on different pathways to induce cell death. The upregulation of BNIP3 upon either chemical or genetic inactivation of G9a might be responsible for synergistic cell death, possibly through an autophagic process. Thus, G9a inhibition may help overcome drug resistance in certain cancer cells.
Although the role of autophagy in tumor formation and progression is still under debate, compelling evidence suggests that either induction or inhibition of autophagy could provide an alternative route towards cancer therapy, especially in cancer cells resistant to chemotherapy-induced apoptosis.11, 12 Gossypol, a natural product isolated from cottonseeds, is reported to be a BH3-mimetic small-molecule inhibitor of BCL2, BCL-xL, BCL-w, and MCL1.31, 32 Both racemic gossypol and (−)-gossypol have anticancer activity against a variety of cancer types and have yielded promising results in ongoing clinical trials.15, 16, 18, 19, 33 Gossypol induces autophagy by blocking the interaction of BCL2/BCL-xL with the proautophagic protein beclin1, releasing beclin1 and triggering an autophagic cascade.17, 34 Both BNIP3 and BNIP3L are implicated in hypoxia-induced autophagy by disrupting the interaction between beclin1 and BCL2/BCL-xL in a similar manner as gossypol.35 It is plausible that the increased BNIP3 and BNIP3L caused by G9a inhibition might reduce the concentration of gossypol required to induce autophagy-related cell death.
There are a few possible mechanisms for the induction of BNIP3 and BNIP3L by G9a inactivation. G9a has been linked to aberrant silencing of several tumor-suppressor genes through H3K9 methylation and DNA CpG methylation.36, 37 The G9a inhibitor BIX01294 reduced repressive chromatin marks at several G9a target genes.38 BNIP3 is downregulated in most pancreatic adenocarcinoma cell lines, including PANC-1, and it may be reactivated by inhibiting gene methylation.39 On the other hand, G9a methylates reptin, a chromatin-remodeling factor under hypoxic conditions, thus negatively regulating the transcription of hypoxia-induced genes, including BNIP3 and BINP3L.40 Further studies need to be conducted to determine whether the activation of BNIP3 and BNIP3L is mediated by direct induction of H3K9 methylation or by indirect activation through reptin, a non-histone substrate of G9a.
Combinatorial drug treatment is widely used in cancer therapeutics to combat single-drug resistance, overcome side effects, and increase sensitivity to therapy. Epigenetic compounds, such as HDAC and DNMT inhibitors, are used for cancer treatment in conjunction with a variety of traditional chemotherapies and radiation therapies.41 Here, we show that HMT inhibitors are capable of synergizing with gossypol to induce cell death in p53-mutant cancer cells, possibly by increasing autophagosome formation. Thus, HMT inhibitors may have promise as sensitizers in apoptosis-resistant cancer cells and are worth further in vivo study.
Materials and Methods
Cell culture and compound treatment
MCF7, HPAC, PANC-1, HeLa, PC-3 (each of which was obtained from ATCC, Manassas, VA, USA), and mCherry-eGFP-LC3 cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco) and 100 U/ml of penicillin–streptomycin (Mediatech, Manassas, VA, USA). hHPNE cells (ATCC) were maintained in the same medium, supplemented with 10 ng/ml human recombinant EGF (Millipore, Billerica, MA, USA) and 750 ng/ml puromycin (Invitrogen, Grand Island, NY, USA). BRD4770 was synthesized as described by Yuan et al.8 Racemic gossypol, UNC0638, HA14-1, rapamycin, tamoxifen, clonidine, carbamazepine, verapamil, and loperamide were purchased from Sigma-Aldrich (St. Louis, MO, USA). ABT-737 was purchased from Selleck Chemicals (Houston, TX, USA). PI3Kg inhibitor was purchased from Echelon Biosciences (Salt Lake City, UT, USA).
Cellular ATP assay
PANC-1 and hHPNE cells were seeded at 1000 cells per well using a Multidrop Combi (Thermo Labsystems, Waltham, MA, USA) in white optical 384-well plates (Corning Life Sciences, Big Flats, NY, USA). After overnight incubation, cells were treated with 2.5 μM BRD4770, and 0.1 μl of a library of 198 bioactive compounds in four doses was added by pin transfer (CyBi-Well Vario, CyBio, Woburn, MA, USA). After incubation for 72 h, medium was removed and 30 μl CellTiter-Glo reagent (Promega, Madison, WI, USA) was added. Luminescence was measured after 10-min incubation using an EnVision plate reader (PerkinElmer, Waltham, MA, USA).
Phenotype microarrays
Cellular metabolism was measured using Phenotype Microarrays (Biolog Inc.) as described previously.13 Briefly, PANC-1 and hHPNE cells were resuspended in a phenol red-free RPMI-1640-based medium depleted of carbon energy sources and containing 2.5 μM BRD4770 and seeded at 30 000 cells per well in four precoated 96-well plates (Biolog Inc.). After 48-h incubation, glucose and Redox Dye Mix MB were added to each well, and cells were incubated in an OmniLog plate reader to assess dye reduction and purple color formation for 24 h.
Small interfering RNA and plasmid transfection
Small interfering RNAs (siRNAs) against GLP (s36392) and G9a (s21469) were obtained from Applied Biosystems (Grand Island, NY, USA). Plasmids encoding human BNIP3 (RC205087) and BNIP3L (RC203315) were obtained from Origene (Rockville, MD, USA). siRNAs (25 nM) and plasmid DNA (0.4 mM) were transfected into PANC-1, mCherry-eGFP-LC3 HeLa, and PC-3 cells using Lipofectamine 2000 reagent (Invitrogen). Transfected cells were cultured for 72 h, followed by mRNA collection for real-time PCR, cell lysis for western blot analysis, and performance of cell-based assays.
Western blotting
Cells were lysed in RIPA buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, protease inhibitor, and phosphatase inhibitor). Total protein was separated by 4–12% SDS-PAGE and transferred to a PVDF membrane using the iBlot system (Invitrogen). Blots were developed using the chemiluminescence detection system SuperSignal (Thermo Fisher Scientific, Waltham, MA, USA), and light emission was captured using Image Station 4000MM Pro (Carestream Health, Rochester, NY, USA). The following antibodies were used for PANC-1 cells, all at 1 : 1000 dilution: G9a rabbit polyclonal antibody (Millipore, no. 07–551), BNIP3 rabbit polyclonal antibody (Millipore, no. AB2999), phospho-p53 (Ser15) mouse monoclonal antibody (mAb) (Cell Signaling Technology, Danvers, MA, USA no. 9287), acetyl-p53 (Lys382) rabbit polyclonal antibody (Cell Signaling, Technology, no. 2525), BCL-xL (54H6) rabbit mAb (Cell Signaling Technology, no. 2764), and LC3B (D11) XP rabbit mAb (Cell Signaling Technology, no. 3868). Secondary horseradish peroxidase-conjugated goat anti-mouse antibody and goat anti-rabbit antibody (Thermo Fisher Scientific) were used at 1 : 10 000 dilution. The following antibodies were used for HeLa cells, all at 1 : 5000 dilution: LC3B rabbit polyclonal antibody (Sigma-Aldrich, L7543), β-actin rabbit mAb (Cell Signaling Technology, no. 4970), and HRP-conjugated anti-rabbit IgG antibody (Cell Signaling Technology, no. 7074).
Gene expression
Total RNA was extracted from treated cells using RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA). Reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems). The Human Autophagy RT2 Profiler PCR array was purchased from SABiosciences (Valencia, CA, USA). qPCR reactions were run in a 384-well format using a LightCycler 480 (Roche, Applied Science, Madison, WI, USA). Primers purchased from Qiagen were GADD45A (QT00014084), MDM2 (QT00056378), GADD45B (QT00018480), BAX (QT00031192), CCNG1 (QT00013916), CDKN1A (QT00062090), CCNG2 (QT00998193), CADD45G (QT00200557), EHMT1 (QT00084602), and EHMT2 (QT00088627).
Microscopy
mCherry-eGFP-LC3 HeLa cells were seeded in black optical 96-well plates (Corning Life Sciences) and either treated with compounds or transfected with plasmids. The cells were then washed with PBS, stained with Hoechst 33342 (Invitrogen), and observed using a fluorescence microscope (Axiovert 200M, Zeiss, Peabody, MA, USA) with × 40 lens.
Synergy calculation
CompuSyn (ComboSyn Inc., Paramus, NJ, USA) was used to calculate the CI of each compound combination according to the method of Chou and Talalay.42, 43
Luciferase reporter assay
MCF7 and PANC-1 cells were treated with BRD4770 or nutlin-3 and reporter plasmids. The activity of p53 was measured following the manuals for Cignal Pathway Reporter Kits (SABiosciences) and Dual-Luciferase Reporter Assay System (Promega).
Affymetrix microassay
PANC-1 cells were treated with DMSO and 10 μM BRD4770 for 6 h; human_U133_plus 2 chip was used. The Human Autophagy RT2 ProfilerTM PCR array was purchased from SABiosciences.
Abbreviations
- HMT:
-
histone methyltransferase
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- BCL2:
-
B-cell lymphoma 2
- BH3:
-
BCL2 homology domain 3
- DMSO:
-
dimethylsulfoxide
- DNMT:
-
DNA methyltransferase
References
Kellen JA . Molecular interrelationships in multidrug resistance. Anticancer Res 1994; 14: 433–435.
Lowe SW, Ruley HE, Jacks T, Housman DE . p53-Dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993; 74: 957–967.
Kristensen LS, Nielsen HM, Hansen LL . Epigenetics and cancer treatment. Eur J Pharmacol 2009; 625: 131–142.
Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 2008; 3: e2037.
Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 2010; 70: 7830–7840.
Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002; 16: 1779–1791.
Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010; 285: 9636–9641.
Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, Adams DJ et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem Biol 2012; 7: 1152–1157.
Lee KM, Yasuda H, Hollingsworth MA, Ouellette MM . Notch 2-positive progenitors with the intrinsic ability to give rise to pancreatic ductal cells. Lab Invest 2005; 85: 1003–1012.
Kroemer G, Marino G, Levine B . Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293.
Kondo Y, Kanzawa T, Sawaya R, Kondo S . The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005; 5: 726–734.
Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis 1996; 17: 1595–1607.
Bochner BR, Siri M, Huang RH, Noble S, Lei XH, Clemons PA et al. Assay of the multiple energy-producing pathways of mammalian cells. PLoS One 2011; 6: e18147.
Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 2011; 7: 566–574.
Liu S, Kulp SK, Sugimoto Y, Jiang J, Chang HL, Dowd MK et al. The (−)-enantiomer of gossypol possesses higher anticancer potency than racemic gossypol in human breast cancer. Anticancer Res 2002; 22: 33–38.
Balakrishnan K, Wierda WG, Keating MJ, Gandhi V . Gossypol, a BH3 mimetic, induces apoptosis in chronic lymphocytic leukemia cells. Blood 2008; 112: 1971–1980.
Lian J, Wu X, He F, Karnak D, Tang W, Meng Y et al. A natural BH3 mimetic induces autophagy in apoptosis-resistant prostate cancer via modulating Bcl-2–Beclin1 interaction at endoplasmic reticulum. Cell Death Differ 2011; 18: 60–71.
Oliver CL, Bauer JA, Wolter KG, Ubell ML, Narayan A, O’Connell KM et al. In vitro effects of the BH3 mimetic, (−)-gossypol, on head and neck squamous cell carcinoma cells. Clin Cancer Res 2004; 10: 7757–7763.
Voss V, Senft C, Lang V, Ronellenfitsch MW, Steinbach JP, Seifert V et al. The pan-Bcl-2 inhibitor (−)-gossypol triggers autophagic cell death in malignant glioma. Mol Cancer Res 2010; 8: 1002–1016.
Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y et al. Molecular cloning and characterization of rat LC3A and LC3B – two novel markers of autophagosome. Biochem Biophys Res Commun 2006; 339: 437–442.
Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4: 151–175.
Fleming A, Noda T, Yoshimori T, Rubinsztein DC . Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol 2011; 7: 9–17.
Tagscherer KE, Fassl A, Campos B, Farhadi M, Kraemer A, Bock BC et al. Apoptosis-based treatment of glioblastomas with ABT-737, a novel small molecule inhibitor of Bcl-2 family proteins. Oncogene 2008; 27: 6646–6656.
van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006; 10: 389–399.
Manero F, Gautier F, Gallenne T, Cauquil N, Gree D, Cartron PF et al. The small organic compound HA14-1 prevents Bcl-2 interaction with Bax to sensitize malignant glioma cells to induction of cell death. Cancer Res 2006; 66: 2757–2764.
Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ 2001; 8: 367–376.
Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K et al. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 2003; 22: 4734–4744.
Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29: 2570–2581.
Kang R, Zeh HJ, Lotze MT, Tang D . The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571–580.
Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S . Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 2004; 64: 4286–4293.
Kitada S, Leone M, Sareth S, Zhai D, Reed JC, Pellecchia M . Discovery, characterization, and structure–activity relationships studies of proapoptotic polyphenols targeting B-cell lymphocyte/leukemia-2 proteins. J Med Chem 2003; 46: 4259–4264.
Etxebarria A, Landeta O, Antonsson B, Basanez G . Regulation of antiapoptotic MCL-1 function by gossypol: mechanistic insights from in vitro reconstituted systems. Biochem Pharmacol 2008; 76: 1563–1576.
Gao P, Bauvy C, Souquere S, Tonelli G, Liu L, Zhu Y et al. The Bcl-2 homology domain 3 mimetic gossypol induces both beclin 1-dependent and beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem 2010; 285: 25570–25581.
Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between beclin 1 and Bcl-2/Bcl-X(L). Autophagy 2007; 3: 374–376.
Zhang J, Ney PA . Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 2009; 16: 939–946.
Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW . 5-Aza-2'-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007; 26: 77–90.
Xin ZH, Ma HL, Wu SY, Dai CH . [Determination of the inhibitory activity of angiotensin-converting enzyme inhibitor captopril by high performance capillary electrophoresis.]. Yao Xue Xue Bao 2003; 38: 843–845.
Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25: 473–481.
Okami J, Simeone DM, Logsdon CD . Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res 2004; 64: 5338–5346.
Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM et al. Negative regulation of hypoxic responses via induced reptin methylation. Mol Cell 2010; 39: 71–85.
Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011; 1: 598–607.
Chou TC, Talalay P . Generalized equations for the analysis of inhibitions of Michaelis–Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur J Biochem 1981; 115: 207–216.
Chou TC . Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006; 58: 621–681.
Acknowledgements
This work was supported by the NIGMS (GM38627 to SL Schreiber) and the NIDDK (DK083048 to BK Wagner). SLS is a Howard Hughes Medical Institute investigator. The content of this paper does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US government.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Yuan, Y., Tang, A., Castoreno, A. et al. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis 4, e690 (2013). https://doi.org/10.1038/cddis.2013.191
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.191
Keywords
This article is cited by
-
Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
BRD4770 inhibits vascular smooth muscle cell proliferation via SUV39H2, but not EHMT2 to protect against neointima formation
Human Cell (2023)
-
Cottonseed extracts regulate gene expression in human colon cancer cells
Scientific Reports (2022)
-
Cottonseed-derived gossypol and ethanol extracts differentially regulate cell viability and VEGF gene expression in mouse macrophages
Scientific Reports (2021)
-
Gossypol decreased cell viability and down-regulated the expression of a number of genes in human colon cancer cells
Scientific Reports (2021)