Abstract
Several advances in recent years have focused increasing attention on the role of the RAF-MEK-ERK1/2 pathway in promoting cell survival. The demonstration that BRAF is a human oncogene mutated at high frequency in melanoma, thyroid and colon cancer has provided a pathophysiological context, whilst the description of potent and highly selective inhibitors of BRAF or MEK has allowed a more informed and rational intervention in both normal and tumour cells. In addition, separate studies have uncovered new mechanisms by which the ERK1/2 pathway can control the activity or abundance of members of the BCL-2 protein family to promote cell survival. It is now apparent that various oncogenes co-opt ERK1/2 signalling to de-regulate these BCL-2 proteins and this contributes to, and even underpins, survival signalling in some tumours. New oncogene-targeted therapies allow direct or indirect inhibition of ERK1/2 signalling and can cause quite striking tumour cell death. In other cases, inhibition of the ERK1/2 pathway may be more effective in combination with other conventional and novel therapeutics. Here, we review recent advances in our understanding of how the ERK1/2 pathway regulates BCL-2 proteins to promote survival, how this is de-regulated in tumour cells and the opportunities this might afford with the use of new targeted therapies.
Similar content being viewed by others
Main
The RAF-MEK-ERK1/2 pathway has often played ‘second fiddle’ to the PI3K pathway in terms of cell survival signalling. Early studies identified and focused on the role of PI3K and the identification of PKB/Akt as an oncogene that prevented apoptosis fuelled interest in the role of the PI3K-PDK1-PKB pathway downstream of survival factors and mutant oncoproteins, notably RAS.1 At the same time, studies defined the importance of the RAF-MEK-ERK1/2 pathway in G1/S cell cycle progression,2 leading to a general perception that the ERK1/2 pathway controlled cell cycle re-entry and the PKB pathway controlled cell survival. However, the emergence of credible evidence of ERK1/2-dependent survival signalling3, 4, 5 made this convenient demarcation untenable and it is now apparent that both pathways can contribute to cell proliferation and cell survival.
The first notable review of ERK1/2 survival signalling stated that ‘perhaps the primary and most defined survival signalling cascade in many cell types involves PI3K and its downstream target Akt or protein kinase B’.6 This may well be the case in ‘normal’ cells but in tumour cells things are a little more complicated. As a result of different oncogene mutations, tumour cells re-model their signalling to acquire key cancer-specific traits7 and the extent to which different pathways are used for survival will depend on the nature of the mutant oncoproteins. In this context, perhaps the most important observation since Ballif and Blenis6 reviewed ERK1/2-dependent survival signalling has been the identification of BRAF as a human oncogene.8 In addition, new mechanisms of ERK1/2-dependent survival signalling have emerged in recent years. In this review, we will describe these new mechanisms, consider how tumours utilize ERK1/2 signalling for aberrant cell survival and discuss the therapeutic opportunities this may afford. We will first start by briefly reviewing how we define ERK1/2-dependent survival signalling.
ERK1/2-Dependent Survival Signalling: Definitions and Tools for Study
Survival factors and many oncoproteins can activate an array of signalling pathways and it is important to define the contribution of each pathway both to fully understand cell survival signalling and to validate individual pathways as therapeutic targets. To deconstruct ERK1/2 survival signalling we can ask whether it is necessary for cell survival response, by the use of selective pathway inhibitors, or sufficient, by the use of activated mutants of RAF or MEK.
Many studies defining the role of the ERK1/2 pathway have used the MEK inhibitors, PD98059 or U0126.6 However, PD98059 is not very potent and has poor solubility and both drugs can partially inhibit MEK5 and thereby inhibit the parallel ERK5 pathway9 so their use requires that the potential involvement of ERK5 is considered as a caveat. The second-generation MEK inhibitors, typified by PD184352 and AZD6244 (ARRY-142886), do not inhibit the ERK5 pathway at physiologically relevant doses and are now the ‘drugs of choice’ for selective inhibition of the ERK1/2 pathway.10, 11, 12 The RAF inhibitor, BAY 43-9006, shows great promise in the clinic but should be used with caution as a research tool as it also inhibits a subset of receptor tyrosine kinases.13 In terms of specificity for the activated, mutant form of BRAF PLX4720 probably represents the most potent and selective option at present.14 It is also desirable to use molecular genetic intervention such as expression of interfering mutants of MEK1/2 or ERK1/2, or the ERK1/2-specific phosphatase DUSP6, which inhibits ERK1/2-dependent signalling without effect on ERK5.15
Constitutively active mutants of RAF or MEK can be transiently or stably expressed in cells to examine their effect on cell survival; these constructs fail to activate ERK5 and their use with a MEK inhibitor can define ERK1/2-dependent survival. However, transient expression of these constructs causes a slow increase in ERK1/2 activation over many hours, making it difficult to define direct and indirect effects. The conditional kinase ΔRAF-1:ER*16 currently represents the best means of rapidly and specifically activating ERK1/2. However, it is still important to differentiate between direct effects of ERK1/2 signalling and indirect effects because of ERK1/2 influencing the activity of other pathways. For example, activation of ΔRAF-1:ER* can protect fibroblasts and epithelial cells against death arising from serum withdrawal or loss of ECM attachment (anoikis);5, 17, 18 however, in MCF-10A cells the protection provided by ΔRAF-1:ER* is because of expression and secretion of EGFR ligands, which act in an autocrine manner to activate PI3K-dependent cell survival.18 In contrast, protection afforded by ΔRAF-1:ER* in other cell systems appears to be a more direct, cell autonomous effect of ERK1/2.4, 5, 17 Clearly, both mechanisms are important but in this review we will focus on cell autonomous mechanisms of cell survival signalling by ERK1/2 and in particular on the regulation of BCL-2 proteins.
The BCL-2 Protein Family
The cell intrinsic apoptosis pathway is regulated by the BCL-2 proteins, which control the release of cytochrome c from mitochondria, triggering the activation of caspases and cell death. The BCL-2 family consists of pro-survival and pro-death proteins that share a number of BCL-2 homology (BH) domains (BH1-4).19 In the pro-survival proteins (such as BCL-2, BCL-xL and MCL-1) the BH1-3 domains form a hydrophobic pocket that can bind the BH3 domains of certain other family members. The multi-domain pro-apoptotic proteins (BAX and BAK) also use BH1-3 to form a hydrophobic pocket and in many viable cells are found in complex with pro-survival BCL-2 proteins. Finally, the BH3-only proteins (BOPs) share only the BH3 domain and are otherwise structurally diverse reflecting their various modes of regulation in response to cell stress or cell survival signals.20 Although the details are beyond the scope of this review, BAX and/or BAK are normally sequestered by pro-survival BCL-2 proteins in viable cells. In response to stress-induced signals, BOPs are either expressed or activated and bind to pro-survival BCL-2 proteins, promoting the release of BAK/BAX, which can then oligomerize, causing mitochondrial outer membrane permeabilization and release of cytochrome c.
ERK1/2-Dependent Regulation of BH3-Only Proteins
Regulation of BIM binding properties and stability
The BIM gene gives rise to three common splice variants; short (BIMS), long (BIML) and extra-long (BIMEL), of which BIMEL is by far the most abundant in most cell types. Among other responses BIM appears to be important in death arising from withdrawal of survival factors, which results in substantial increases in BIMEL expression. Cytokine withdrawal leads to inactivation of PKB, de-phosphorylation and nuclear entry of FOXO3A, which can then promote BIM expression.21 It has been known for several years that ERK1/2 activation can repress BIM mRNA levels.17 Recently, a potential mechanism to account for this has emerged with the demonstration that ERK1/2 can phosphorylate FOXO3A, targeting it for proteasomal degradation;22 thus both ERK1/2 and PKB can inactivate FOXO3A by different mechanisms to repress BIM (Figure 1). In addition to regulation of transcription, BIMEL is phosphorylated at multiple sites in response to selective activation of the ERK1/2 pathway17 and this has the effect of promoting the ubiquitination and proteasome-dependent turnover of BIMEL23 (Figure 1). BIMEL is phosphorylated directly by ERK1/2 at up to four different sites;24, 25, 26 the exon unique to BIMEL encodes three ERK1/2 phosphorylation sites and a DEF-type ERK1/2 docking domain.27, 28 Of these three Ser69 (Ser65 in mouse and rat) appears to be a key signal for BIMEL turnover as S69A or S69G mutants are defective in turnover, accumulate to higher levels than wild-type protein, and may exhibit enhanced toxicity.24, 29, 30
Regulation of BIM binding properties and stability by ERK1/2. The pro-apoptotic BH3-only protein BIM is expressed de novo following cytokine withdrawal and binds to pro-survival proteins such as MCL-1, thereby releasing BAX or BAK to promote cell death. ERK1/2-catalysed phosphorylation of FOXO3A promotes its proteasomal degradation, thereby preventing FOXO3A-dependent expression of BIM. In addition, BIMEL, the most abundant BIM splice variant, is phosphorylated directly by ERK1/2 on at least three different sites. This promotes the dissociation of BIMEL from pro-survival proteins and targets BIMEL for poly-ubiquitination through the 26S proteasome. See text for details
More recently, it has been demonstrated that ERK1/2 activation can inhibit the binding of BIMEL to BCL-2 family proteins such as BCL-xL and MCL-1.25, 31 In serum-starved cells newly synthesized BIMEL associates with BCL-xL and MCL-1 but re-stimulation of cells with growth factors promotes the rapid dissociation of BIMEL:BCL-xL or BIMEL:MCL-1 complexes and activation of ERK1/2 is necessary and sufficient for this25 (Figure 1). As BIMEL needs to engage with pro-survival proteins to kill cells this represents an additional survival mechanism. Furthermore, BIMEL mutants with impaired binding to MCL-1 or BCL-xL exhibit accelerated turnover, suggesting that ERK1/2-dependent dissociation of BIMEL from pro-survival proteins may be the first step in its proteasomal degradation.25, 31 The precise mechanism of degradation of BIMEL is unclear. It has been suggested that CBL is the E3 ligase responsible for poly-ubiquitination of BIMEL;32 however, others fail to detect binding of BIMEL to CBL and BIMEL is turned over normally in CBL−/− fibroblasts arguing against this.33
Regulation of BAD binding properties and stability
BAD is inhibited by phosphorylation at three distinct sites (Figure 2): Ser112, catalysed by RSK; Ser136, catalysed by PKB and Ser155 catalysed by PKA. Early studies showed that growth factor-stimulated phosphorylation of Ser112 was inhibited by MEK inhibitors34, 35 although Ser136 phosphorylation was inhibited by PI3K inhibitors.36, 37 RSK was subsequently shown to be the kinase responsible for phosphorylation of Ser112.38, 39 Phosphorylation by RSK or PKB inhibits BAD by facilitating its binding to 14-3-3 proteins although PKA-dependent phosphorylation of Ser155 within the BAD BH3 domain blocks interactions with pro-survival proteins, causing BAD to dissociate from the mitochondria.40, 41, 42 These three phosphorylation events may cooperate; for example, the increased binding of BAD to 14-3-3 arising from phosphorylation of Ser112 and Ser136 may facilitate phosphorylation of Ser155 by PKA42 (Figure 2). MSK1, a RSK-related protein kinase that is activated by ERK1/2 or p38-dependent phosphorylation, can also promote BAD phosphorylation at Ser112, providing a further ERK1/2-dependent input into BAD inhibition.43
Regulation of BAD binding properties and stability by ERK1/2 signalling. In cells deprived of cytokines or treated with PI3K and MEK inhibitors BAD is de-phosphorylated and binds to pro-survival proteins such as BCL-xL to promote cell death. In contrast, cytokines promote the phosphorylation of BAD, allowing it to be sequestered by 14-3-3 proteins as a survival mechanism. PKB phosphorylates BAD at Ser136 whereas RSK, a direct substrate of ERK1/2, phosphorylates BAD at Ser112. This dual phosphorylation may in turn facilitate phopsphorylation of Ser155 by cAMP-dependent PKA. Another ERK1/2-dependent kinase, MSK, has also been implicated as a possible Ser112 kinase. In addition, it has recently been shown that activation of RAF promotes the ERK1/2- and RSK-dependent poly-ubiquitination and proteasomal degradation of BAD; this is dependent upon phosphorylation of Ser112. See text for details
Few studies have reported significant changes in BAD abundance. However, it has recently been shown that activation of the ERK1/2 pathway promotes the ubiquitination and proteasome-dependent turnover of BAD that requires RSK-dependent phosphorylation of Ser112.44 This provides the first evidence that the ERK1/2 pathway can promote turnover of BAD (Figure 2) and raises several questions. First, the identification of the relevant E3 ligase for BAD will be of great interest. Second, what contribution will BAD stabilization make to proteasome inhibitor-induced cell death? Finally, is the ERK1/2-induced turnover of BAD cell type-specific, because in a previous study of ERK1/2-dependent BIMEL turnover BAD was actually used as a negative control that did not turnover in response to ERK1/2 activation.23 Such issues should provide a greater insight into the physiological role of BAD protein turnover in ERK1/2 survival signaling.
Regulation of BMF transcription/mRNA stability
Non-transformed MCF-10A human mammary epithelial cells undergo apoptosis when detached from their ECM attachment (so called anoikis). This is accompanied by an increase in BMF mRNA levels, which is also observed in a variety of other epithelial cells, and knockdown of BMF inhibits cell death in suspension.45 Interestingly, expression of activated mutants of HRAS, MEK2, PI3K or PKB protected cells from anoikis and also caused a substantial reduction in BMF mRNA levels. Thus, both the ERK and PKB pathways can repress transcription of BMF or reduce BMF mRNA stability but the underlying mechanism is unclear.
ERK1/2-Dependent Regulation of Pro-Survival BCL-2 Proteins
There is abundant evidence that survival factors can use the ERK1/2 pathway to increase the expression of several pro-survival BCL-2 proteins, notably BCL-2, BCL-xL and MCL-1, by promoting de novo gene expression in a variety of cell types; for example, MEK inhibition caused a decrease in BCL-2, BCL-xL and MCL-1 and apoptosis in pancreatic cancer cells.46 There is a prominent role for CREB in mediating the ERK1/2-dependent expression of these genes by virtue of the fact that they all have CREB-binding sites in their 5′ regulatory regions and the ERK1/2-dependent kinases RSK or MSK can phosphorylate and activate CREB; this has been reviewed previously.6
Regulation of MCL-1 protein stability
MCL-1 expression is frequently elevated in tumours and tumour cell lines and is associated with poor prognosis and drug resistance.47 Among the pro-survival BCL-2 proteins a reduction in MCL-1 abundance is most frequently observed upon inhibition of the ERK1/2 pathway and this partly reflects direct ERK1/2-dependent regulation of MCL-1. The MCL-1 protein exhibits a relatively short half-life and this is attributed to the presence of a PEST domain residing in the N-terminal extension preceding the BH domains; indeed, MCL-1 turnover is regulated by phosphorylation at two sites within this PEST domain. Activation of ERK1/2 stabilizes MCL-1 and this is achieved through the direct ERK1/2-dependent phosphorylation of Thr163 within the PEST domain48 (Figure 3). This may account for instances in which MCL-1 overexpression is correlated with hyper-activation of the ERK1/2 pathway. However, it has also been reported that phosphorylation at Thr163 ‘primes’ MCL-1 for phosphorylation at Ser159 by GSK3β, which apparently enhances MCL-1 poly-ubiquitination and degradation49 (Figure 3).
Regulation of MCL-1 stability by ERK1/2 signalling. Withdrawal of cytokines or inhibition of the ERK1/2 pathway promotes the proteasome-dependent degradation of MCL-1 whereas activation of ERK1/2 inhibits the degradation of MCL-1. ERK1/2 phosphorylates MCL-1 directly at Thr163 within the PEST domain and this stabilizes MCL-1. However, phosphorylation at Thr163 may also ‘prime’ MCL-1 for phosphorylation at Ser159 by GSK3β thereby promoting its turnover. GSK3β is in turn inhibited by both RSK and PKB so that there is a fine interplay between ERK1/2-RSK, PKB and GSK3β in the control of MCL-1 protein levels. The only E3 ubiquitin ligase known to facilitate poly-ubiquitination of MCL-1 is MULE, which binds through a BH3 domain to the hydrophobic groove of MCL-1. It is not known how or indeed if MCL-1 phosphorylation influences MULE binding or whether there is an additional E3 ligase which links MCL-1 phosphorylation and turnover
This reveals a very fine interplay between ERK1/2-dependent stabilization and GSK3β-dependent turnover in the control of MCL-1 protein levels and raises some important questions. First, the only E3 ligase known to catalyse the poly-ubiquitination of MCL-1 is MULE, which contains a BH3-domain through which it binds to MCL-1.50 However, it is presently unclear how phosphorylation at Thr163 or Ser159 within the PEST domain is linked to dissociation or recruitment of MULE to the BH3 domain-binding groove. Second, as ERK1/2 can activate RSK, which in turn can phosphorylate and inhibit GSK3β,51 does RSK activation facilitate ERK1/2-dependent MCL-1 stabilization by inhibiting GSK3β? Finally, how does activation of PKB interface with this ERK/GSK3β regulatory pairing? It is well known that PKB can phosphorylate and inhibit GSK3β and so activation of the PKB pathway should stabilize MCL-1. Similarly, inhibition of PKB, which activates GSK3β, results in downregulation of MCL-1.52 However, if GSK3β-dependent phosphorylation of Ser159 is dependent upon prior phosphorylation by ERK1/2 at Thr163 then turnover of MCL-1 following PI3K/PKB inhibition (a therapeutically desirable event in cancer therapy) may be dependent upon the presence of an active ERK1/2 pathway and prior phosphorylation of Thr163. These latter predictions are readily testable and underscore the complex interplay between the ERK and PI3K-PKB pathways (Figure 3).
Oncogene Addiction and Evolved Dependency upon the ERK1/2 Pathway
Clearly the ERK1/2 pathway can regulate several members of the BCL-2 protein family to achieve cell survival. However, many of these proteins (e.g., BIM, BAD, MCL-1, and so on) are also controlled by other oncogene-regulated signalling pathways, notably the PI3K-PKB pathway; indeed, the use of such redundant mechanisms is the rule rather than the exception. So, if we want to inhibit one of these pathways in the hope of tipping the balance in favour of tumour cell death, which one is more important? Of course, one can determine this empirically but the individual repertoire of mutations that each tumour possesses may also be informative.
During its lifetime a tumour cell accumulates dozens of mutations; some are ‘drivers’ which promote the genesis and maintenance of the tumour whereas others are simply ‘passengers’ and confer no selective advantage.53 The concept of ‘oncogene addiction’54 posits that tumour cells remain dependent upon these driver mutations to maintain their malignant state. Loss of these mutations, or inhibition of the pathways they control, leads to loss of one or more of the key acquired traits of the cancer cell7 leading to loss of tumorigenic potential and/or tumour regression. For example, deletion of the activated KRAS allele in the HCT116 and DLD-1 human colorectal cancer cell lines result in clones that are no longer tumorigenic.55 Despite the fact that these cells have accumulated additional genetic alterations they remain dependent upon the activated KRAS allele, which can be viewed as a foundation of the tumour.
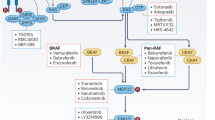
Arising from these observations is an appreciation that tumour cells evolve to be dependent upon the signal pathways activated by their driver oncogenes. For example, if a tumour cell does not have a mutation that activates the ERK1/2 pathway (and so exhibits a low level of ERK1/2 activation) it will make little use of this pathway to protect it from insults but will evolve to be dependent upon other pathways that are activated, such as the PI3K pathway. On the other hand, a tumour cell with a BRAF mutation (frequently an early event) will use ERK1/2 signalling to overcome the selection barriers for survival and will evolve to be dependent upon this pathway. The degree of dependency on ERK1/2 signalling may be modulated by coincident mutations in other pathways. So for example, a tumour cell with a BRAF mutation may be very dependent upon the ERK1/2 pathway whereas that with a KRAS or EGFR mutation may be less so, as these oncoproteins activate multiple effector pathways that may influence cell survival (Figure 4). Here, we discuss examples of tumour cells that are addicted to oncoproteins for cell survival and the contribution that the ERK1/2 pathway makes in regulating BCL-2 proteins. In principle, inactivation of ERK1/2 signalling can increase/activate pro-apoptotic proteins or decrease expression of survival proteins; in practice, both are observed in tumour cells and drug-induced death represents a composite of these.
Oncogene addiction and signal pathway redundancy. BRAF is thought to signal predominantly through the MEK1/2-ERK1/2 pathway (shown in black in left panel) although there is evidence for MEK-independent signalling by RAF isoforms (represented by ?). Consequently a tumour cell with a BRAF mutation will use ERK1/2 signalling to overcome the selection barriers for survival and will evolve to be dependent upon or addicted to this pathway. The degree of dependency on ERK1/2 signalling may be modulated by coincident mutations in other pathways. So for example, a tumour cell with a BRAF mutation may be very dependent upon the ERK1/2 pathway whereas that with a KRAS mutation (middle panel) or an EGFR mutation (right panel) may be less so as these oncoproteins activate other effector pathways that may influence cell survival by regulating common death effectors such as BIM or BAD or survival proteins such as MCL-1. Consequently, a MEK inhibitor (highlighted with a star) may promote a significant pro-apoptotic response in cells with a BRAF mutation and may sensitize such cells to other stresses. In contrast, cells with a KRAS of EGFR mutation may use other pathways to repress BIM and BAD or stabilize MCL-1 and so may be less sensitive to RAF or MEK inhibition
Tumour cells with BRAF600E
BRAF mutations are common in melanoma, thyroid and colorectal cancer8 and cell lines derived from these tumours typically exhibit high ERK1/2 activity. Such cells lines are frequently addicted to ERK1/2 signalling; for example, BRAF600E-positive melanoma cells undergo a G1 arrest and apoptosis when BRAF expression is silenced or the pathway is inhibited even when cells are maintained in complete medium.56 A consequence of this addiction to ERK1/2 signalling is that inhibition of the pathway frequently sensitizes tumour cells to insults they are otherwise relatively resistant to. This is particularly evident in the case of serum withdrawal; for example, BRAF600E-positive melanoma and colorectal cancer cells appear to be addicted to the ERK1/2 pathway for growth factor-independent survival as they undergo cell death when subjected to serum withdrawal combined with MEK inhibition.57, 58 Inhibition of MEK in invasive melanoma cells sensitizes them to anoikis59 whereas papillary thyroid cancer cell lines may or may not undergo apoptosis upon inhibition of the ERK1/2 pathway.60, 61
It is now clear from several studies that BIM is a major target of ERK1/2-dependent survival signalling in BRAF600E-positive tumour cells. In melanoma cell lines, inhibition of the ERK1/2 pathway with PD184352 (CI-1040) synergizes with withdrawal of serum to promote apoptosis; under these conditions there is a strong increase in BIM expression and RNAi-mediated knockdown of BIM confers partial protection.57 Similarly, serum withdrawal and MEK inhibition by U0126 or AZD6244 gives rise to a strong increase in BIM expression in BRAF600E-positive colorectal cancer cell lines and death under these conditions is significantly reduced by siRNA-mediated ablation of BIM.58 Indeed, the clinical candidate AZD6244 caused a striking increase in BIMEL expression in COLO205 cells58 and promoted regression of COLO205 xenografts in vivo.12 Both of these studies reveal that BIMEL is the major form of BIM that is subject to regulation by MEK inhibitors and demonstrate that tumour cells with BRAF600E exhibit a strong, constitutive signal for BIMEL degradation through the proteasome.57, 58 They also reveal some important details: first, inhibition of MEK alone in complete medium can cause an increase in BIM expression but relatively little cell death;57 second, even complete ablation of BIM by siRNA affords only 60% protection against death arising from serum withdrawal and MEK inhibition.58 Both these observations suggest that either BIM expression must reach a certain threshold to induce death or that BIM acts in concert with another BH3-only protein. Indeed, studies have suggested that ERK1/2 signalling protects melanoma cells from anoikis by inhibiting both BAD and BIM.62, 63 Inhibition of ERK1/2 signalling increased the abundance of both BIM and BAD and their combined knockdown strongly inhibited death arising from BRAF knockdown.63 In a separate study, melanoma cell lines with high levels of ERK1/2 signalling underwent apoptosis when treated with U0126 and the increase in BIM and PUMA and the loss of MCL-1 appeared to play the critical role in cell death.64
ERK1/2 signalling can also provide protection against chemotherapeutic cytotoxic drugs. In BRAFV600E-positive melanoma cells ERK1/2 signalling protects against apoptosis induced by cisplatin, actinomycin D or daunorubicin.65 Treatment with UO126 increased BIM expression, decreased BAD phosphorylation and, surprisingly, reduced NOXA expression. Knockdown of either BIM, BAD or NOXA separately had little effect on apoptosis but triple knockdown did afford partial protection;65 whether inhibition of ERK1/2 signalling (which activates BIM and BAD) can synergize with DNA-damaging agents (which work in part through NOXA and PUMA) and which BOPs are involved in such synergy, was not addressed. BIM is required for paclitaxel-induced apoptosis in baby mouse kidney epithelial cells and activation of the ERK1/2 pathway by expression of mutant HRAS or mutant CRAF reduces sensitivity to paclitaxel by promoting BIMEL phosphorylation and turnover.66 However, it remains to be seen if the combination of paclitaxel and RAF or MEK inhibition can synergize to promote cell death in human tumour cells with BRAF600E and whether this is dependent upon BIM. Regardless, these examples reveal the extent to which tumour cells with BRAF mutations can evolve to be dependent upon ERK1/2 signalling for normal survival and against various drug-induced insults. It also indicates that the BCL-2 protein family is recruited to initiate death when the ERK1/2 pathway is inhibited, suggesting that inhibitors of the ERK1/2 pathway may show promise alone or in combination with other therapies in tumours with BRAF600E
Tumour cells with RAS mutations
In contrast to BRAF600E mutant RAS oncoproteins have more options for survival signalling because they activate multiple effector pathways (Figure 4). Although the precise role of these various effectors in RAS-dependent survival signalling and tumorigenesis is not yet known, it is clear that some, notably PI3K, are clearly required for RAS-induced tumorigenesis in mouse models.67 Inhibition of the ERK1/2 pathway is often associated with a G1 cell cycle arrest in cells with RAS mutations68 but there are clearly instances in which ERK1/2 inhibition promotes cell death. For example, pancreatic cancer cells with KRAS mutations may undergo cell cycle arrest69 or cell death46 upon inhibition of the ERK1/2 pathway. In HCT116 colorectal cancer cells, which harbour a KRAS mutation, MEK inhibitors promote a G1 cell cycle arrest followed by a delayed caspase-dependent cell death; although the precise mechanism is unclear, such treatments cause a striking increase in BIM expression (K Balmanno and S Cook, unpublished observations). Indeed, even in melanoma cell lines with NRAS mutations, MEK inhibition invariably leads to an increase in BIM expression65 but this is not always accompanied by cell death indicating that increased BIM expression alone may not be sufficient for cell death. Clearly the use of multiple effector pathways downstream of RAS means that alternative pathways can substitute for ERK in maintaining cell survival. This situation is made more complex by the recognition that the same RAS mutant can activate different pathways to different extents, even in the same tumour types. Analysis of colorectal cancer cell lines with the same KRAS alleles has revealed striking differences in the activation of ERK1/2 or PKB, with some cell lines exhibiting very low levels of ERK1/2 activity and others very high70, 71 (K Balmanno and S Cook, unpublished observations). So death arising from MEK inhibition may be determined by the degree to which RAS mutants couple to ERK signalling (pathway addiction) and the presence of mutations on other pathways such as PI3K which offset this. The activation of multiple effector pathways is exacerbated still further in the case of oncoproteins such as EGFR or BCR/ABL (Figure 4).
EGFR in lung cancer
Overexpression of EGFR is seen in many tumours of epithelial origin, including cancers of the lung, breast, head and neck and bladder, and is associated with a poor prognosis. Some non-small cell lung cancer cells exhibit somatic point mutations in EGFR and are remarkably sensitive to the EGFR-selective tyrosine kinase inhibitors (TKIs), gefitinib or erlotinib, which inhibit the otherwise constitutive activation of the ERK1/2 and PKB pathways. Treatment of sensitive NSCLC cells with gefitinib72, 73 or erlotinib74, 75 increased the expression of BIM in all studies and also reduced the expression of MCL-1.74 The reduction in MCL-1 might contribute to apoptosis but was not consistently observed in drug-sensitive cell lines. It is also likely that other BCL-2 family proteins, such as BAD and PUMA, may be involved in the induction of apoptosis in NSCLC by erlotinib.74, 75 Erlotinib can also inhibit growth of lung cancer xenografts in nude mice and this is associated with increased BIM expression.74 In all these studies upregulation of BIM was invariably a response to EGFR inhibition and was required for maximal apoptosis. Finally, secondary mutations in the EGFR that are seen in relapsing patients with acquired resistance prevent the increased BIM expression and cell death in response to gefitinib.72
These studies reveal the remarkable extent to which certain NSCLC cells are addicted to signalling by the EGFR and the importance of BIM and potentially BAD in cell death arising from EGFR inhibition. What is less clear is the role of loss of ERK1/2 signalling in the erlotinib- or gefitinib-induced death. Even though MEK inhibitors and erlotinib or gefitinib induced comparable levels of BIM expression and BIMEL dephosphorylation, inhibition of the ERK1/2 pathway alone did not induce significant levels of apoptosis in TKI-sensitive NSCLC cells.73 Indeed, a study using gefitinib-resistant NSCLC cells has previously suggested that complete inhibition of both the ERK1/2 and PI3K pathways is required to induce apoptosis in these cells.76 Thus, the ERK1/2 pathway forms part of a cell survival signalling network downstream of EGFR in NSCLC, but its inhibition alone may not be sufficient for cell death. Similarly, this implies that the expression and de-phosphorylation of BIMEL arising from MEK inhibition may not be sufficient for cell death in these cells. Indeed, enhanced NSCLC cell killing is observed when targeted TKIs are combined with BH3 mimetic therapeutics such as ABT-73773 suggesting that multiple pro-survival proteins need to be inhibited for optimal therapeutic cell killing or the increase in BIM expression does not reach a sufficient threshold for cell death.
BCR/ABL in chronic myelogenous leukaemia
In chronic myelogenous leukaemia the BCR/ABL oncoprotein activates a number of signalling cascades including the RAS-RAF-MEK-ERK1/2 pathway, the PI3K pathway, STAT5 and NF-κB.77 Activation of these pathways leads to growth factor-independent growth and survival, which can be overcome by targeted TKI therapies against the BCR/ABL tyrosine kinase such as imatinib and dasatinib. BCR/ABL signalling increases the expression of BCL-XL, MCL-1 and BCL-2, which may have a role in protecting chronic myelogenous leukaemia (CML) cells from apoptosis.78, 79, 80, 81 Certainly imatinib treatment causes a striking loss of MCL-1 in CML cells and knockdown of MCL-1 reduces cell viability.80 There is also a prominent role for BIM in death arising from BCR/ABL inhibition. Expression of BIM is repressed in blast cells from mouse models of CML and from patients.81, 82 Treatment of these cells with imatinib increased BIM expression and cells died in a BIM-dependent fashion.81, 82 In BCR/ABL-positive K562 cells BIM knockdown did not provide complete protection against imatinib-induced apoptosis but knockdown of BIM and BAD together almost completely protected BCR/ABL-transformed murine progenitor cells from imatinib-induced apoptosis suggesting that BAD and BIM play a role in TKI-induced apoptosis in CML.83 Furthermore, the BH3 mimetic ABT-737 could enhance cell killing induced by imatinib and could even overcome resistance associated with loss of BIM and BAD.83
In haematopoietic progenitor cells IL-3 represses BIM expression primarily through the ERK1/2 pathway but also requires additional signals for cell survival.84 The same seems to apply for BCR/ABL as treatment of BCR/ABL-transformed Ba/F3 cells with MEK inhibitors induced BIM expression to the same degree as that observed with imatinib, whereas PI3K inhibition had little effect.82 In addition, both imatinib and MEK inhibitors induce a similar loss of MCL-1 in CML cells.80 Despite this, as with EGFR in NSCLC, inhibition of ERK1/2 signalling alone with PD184352 caused little cell death in BCR/ABL-positive leukaemic cells85 indicating that BCR/ABL uses multiple pathways to ensure cell survival. The combination of the dual Src/Abl inhibitor dasatinib and PD184352 gave a marked enhancement of apoptosis compared with either drug given alone, including in an imatinib-resistant clone of K562 cells; death under these conditions appears to be due to expression of BIM and the loss of MCL-1.85 These results highlight the importance of coordinated changes in expression and activity of BCL-2 proteins in the cytotoxic response to TKIs and the potential of augmenting this by co-administration of inhibitors of the ERK1/2 pathway or BH3 mimetics.
Conclusions
The last 5–6 years has seen the discovery of a variety of new mechanisms by which the ERK1/2 pathway can regulate BCL-2 proteins to promote cell survival and a growing appreciation of the extent to which these mechanisms are co-opted by oncoproteins in tumour cells. In many cases both the ERK1/2 and PI3K-PKB pathways regulate a common set of cell death regulators (BIM, BAD, BMF, MCL-1, BCL-xL, BCL-2) indicating a significant degree of redundancy in survival signalling. Such a redundancy should come as no surprise but represents a challenge when seeking to inhibit selected pathways for therapeutic effect in cancer. The importance of ERK1/2 in survival signalling in cancer will be determined by the remodelling of survival signalling in cancer cells; this will differ for different oncogenes so the context is key. For example, cancers with BRAF mutations may evolve to be addicted to the ERK1/2 pathway so RAF or MEK inhibition may represent a potential therapeutic strategy. In tumours with RAS or tyrosine kinase mutations, ERK1/2 may be just one arm of a broad survival signalling network (Figure 4). In such cases RAF or MEK inhibition alone may have limited efficacy but may be more effective when combined with TKIs, PI3K or PKB inhibitors. Indeed, there are clear examples in which inhibition of ERK1/2 signalling can cooperate with targeted TKIs to de-regulate BCL-2 proteins and promote tumour cell death. In instances where RAF or MEK inhibitors do induce tumour cell death there is good evidence to suggest that this proceeds through the BCL-2 proteins. In this context, studies on the combination of RAF or MEK inhibitors with BH3 mimetics such as ABT-737 in tumours and tumour cell lines with high levels of ERK1/2 activity are perhaps overdue. Finally, although inhibitors of BRAF or MEK catalytic activity may hold promise, it is worth remembering that CRAF can provide survival signals that do not depend on its catalytic activity86 and these may be co-opted by RAS or certain tumour-derived BRAF mutants with reduced kinase activity.87 Understanding potential MEK-independent effects of BRAF or CRAF signalling may prove to be very important.
Although this review has focused on the regulation of BCL-2 proteins it is clear that ERK1/2 signalling can also promote cell survival by other mechanisms. For example, ERK1/2 activation can inhibit apoptotic signalling by Fas, TNF, and TRAIL receptors88 suggesting that ERK1/2 inhibition might be effective in combination with recombinant ligands or agonistic mAbs for these receptors in tumours with de-regulated ERK1/2 signalling. What is clear is that the ERK1/2 pathway is no longer ‘second fiddle’ in survival signalling; rather, it is an important component of cell survival signalling networks and in certain tumour types may ‘lead the orchestra’.
Abbreviations
- BAD:
-
BCL-xL/BCL-2-associated death promoter
- BCL-2:
-
breakpoint cluster-2
- BCL-xL:
-
Basal cell lymphoma-extra large
- BCR/ABL:
-
Breakpoint cluster region/Abelson murine leukaemia viral oncogene
- BH:
-
BCL-2 homology
- BIM:
-
BCL-2 interacting mediator of cell death
- BMF:
-
BCL-2 modifying factor
- BOP:
-
BH3-only protein
- CBL:
-
Casitas B-lineage lymphoma oncogene
- CML:
-
chronic myelogenous leukaemia
- CREB:
-
cAMP-response element binding protein
- DUSP:
-
Dual-specificity phosphatase
- EGFR:
-
epidermal growth factor receptor
- ERK:
-
Extracellular signal-regulated kinase
- FOXO:
-
Forkhead box
- GSK3:
-
glycogen synthase kinase 3
- IL-3:
-
interleukin-3
- JNK:
-
c-Jun N-terminal kinase
- MAPK:
-
mitogen-activated protein kinase
- MCL-1:
-
myeloid cell leukaemia 1
- MEK:
-
MAPK or ERK Kinase
- MSK:
-
mitogen- and stress-activated protein kinase 1
- NFκB:
-
nuclear factor kappa B
- NOXA:
-
'damage'
- NSCLC:
-
non-small cell lung cancer
- PDK1:
-
phosphoinositide-dependent kinase 1
- PEST:
-
proline-, glutamic-, serine- and threonine-rich
- PI3K:
-
phosphoinositide 3′-OH kinase
- PKA:
-
cAMP-dependent protein kinase A
- PKB:
-
Protein Kinase B (also known as Akt)
- PUMA:
-
p53-upregulated modulator of apoptosis
- RSK:
-
Ribosomal protein S6 kinase
- STAT:
-
signal transducer and activator of transcription
- TKI:
-
Tyrosine kinase inhibitor
References
Downward J . Ras signalling and apoptosis. Curr Opin Genet Dev 1998; 8: 49–54.
Lavoie JN, Rivard N, L'Allemain G, Pouysségur J . A temporal and biochemical link between growth factor-activated MAP kinases, cyclin D1 induction and cell cycle entry. Prog Cell Cycle Res 1996; 2: 49–58.
Kinoshita T, Shirouzu M, Kamiya A, Hashimoto K, Yokoyama S, Miyajima A . Raf/MAPK and rapamycin-sensitive pathways mediate the anti-apoptotic function of p21Ras in IL-3-dependent hematopoietic cells. Oncogene 1997; 15: 619–627.
Erhardt P, Schremser EJ, Cooper GM . B-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathway. Mol Cell Biol 1999; 19: 5308–5315.
Le Gall M, Chambard JC, Breittmayer JP, Grall D, Pouysségur J, Van Obberghen-Schilling E . The p42/p44 MAP kinase pathway prevents apoptosis induced by anchorage and serum removal. Mol Biol Cell 2000; 11: 1103–1112.
Ballif BA, Blenis J . Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Diff 2001; 12: 397–408.
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417: 949–954.
Kamakura S, Moriguchi T, Nishida E . Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J Biol Chem 1999; 274: 26563–26571.
Squires MS, Nixon PM, Cook SJ . Cell-cycle arrest by PD184352 requires inhibition of extracellular signal-regulated kinases (ERK) 1/2 but not ERK5/BMK1. Biochem J 2002; 366: 673–680.
Davies SP, Reddy H, Caivano M, Cohen P . Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 2000; 351: 95–105.
Davies BR, Logie A, McKay JS, Martin P, Steele S, Jenkins R et al. AZD6244 (ARRY-142886), a potent inhibitor of MEK1/2: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship and potential for combination in preclinical models. Mol Cancer Ther 2007; 6: 2209–2219.
Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 2004; 64: 7099–7109.
Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 2008; 105: 3041–3046.
Arkell RS, Dickinson RJ, Squires M, Hayat S, Keyse SM, Cook SJ . DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5. Cell Signal 2008; 20: 836–843.
Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M . Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol 1997; 17: 5598–5611.
Weston CR, Balmanno K, Chalmers C, Hadfield K, Molton SA, Ley R et al. Activation of ERK1/2 by ΔRaf-1:ER* represses Bim expression independently of the JNK or PI3K pathways. Oncogene 2003; 22: 1281–1293.
Schulze A, Lehmann K, Jefferies HB, McMahon M, Downward J . Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev 2001; 15: 981–994.
Adams JM, Cory S . The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007; 26: 1324–1337.
Puthalakath H, Strasser A . Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 2002; 9: 505–512.
Fu Z, Tindall DJ . FOXOs, cancer and regulation of apoptosis. Oncogene 2008; 27: 2312–2319.
Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 2008; 10: 138–148.
Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ . Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 2003; 278: 18811–18816.
Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ . Extracellular signal-regulated kinases 1/2 are serum-stimulated ‘BimEL kinases’ that bind to the BH3-only protein Bim(EL) causing its phosphorylation and turnover. J Biol Chem 2004; 279: 8837–8847.
Ewings KE, Hadfield-Moorhouse K, Wiggins CM, Wickenden JA, Balmanno K, Gilley R et al. ERK1/2-dependent phosphorylation of BimEL promotes its rapid dissociation from Mcl-1 and Bcl-xL. EMBO J 2007; 26: 2856–2867.
Hübner A, Barrett T, Flavell RA, Davis RJ . Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 2008; 30: 415–425.
Ley R, Hadfield K, Howes E, Cook SJ . Identification of a DEF-type docking domain for extracellular signal-regulated kinases 1/2 that directs phosphorylation and turnover of the BH3-only protein BimEL. J Biol Chem 2005; 280: 17657–17663.
Ley R, Ewings KE, Hadfield K, Cook SJ . Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ 2005; 12: 1008–1014.
Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 2003; 22: 6785–6793.
Marani M, Hancock D, Lopes R, Tenev T, Downward J, Lemoine NR . Role of Bim in the survival pathway induced by Raf in epithelial cells. Oncogene 2004; 23: 2431–2441.
Ewings KE, Wiggins CM, Cook SJ . Bim and the pro-survival Bcl-2 proteins: opposites attract, ERK repels. Cell Cycle 2007; 6: 2236–2240.
Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J 2003; 22: 6653–6664.
Wiggins CM, Band H, Cook SJ . c-Cbl is not required for ERK1/2-dependent degradation of BimEL. Cell Signal 2007; 19: 2605–2611.
Fang X, Yu S, Eder A, Mao M, Bast Jr RC, Boyd D et al. Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene 1999; 18: 6635–6640.
Scheid MP, Schubert KM, Duronio V . Regulation of bad phosphorylation and association with Bcl-x(L) by the MAPK/Erk kinase. J Biol Chem 1999; 274: 31108–31113.
Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997; 91: 231–241.
del Peso L, González-García M, Page C, Herrera R, Nuñez G . Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 1997; 278: 687–689.
Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME . Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999; 286: 1358–1362.
Shimamura A, Ballif BA, Richards SA, Blenis J . Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol 2000; 10: 127–135.
Lizcano JM, Morrice N, Cohen P . Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J 2000; 349: 547–557.
Virdee K, Parone PA, Tolkovsky AM . Phosphorylation of the pro-apoptotic protein BAD on serine 155, a novel site, contributes to cell survival. Curr Biol 2000; 10: 1151–1154.
Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell 2000; 6: 41–51.
She QB, Ma WY, Zhong S, Dong Z . Activation of JNK1, RSK2, and MSK1 is involved in serine 112 phosphorylation of Bad by ultraviolet B radiation. J Biol Chem 2002; 277: 24039–24048.
Fueller J, Becker M, Sienerth AR, Fischer A, Hotz C, Galmiche A . C-RAF activation promotes BAD poly-ubiquitylation and turn-over by the proteasome. Biochem Biophys Res Commun 2008; 370: 552–556.
Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci USA 2007; 104: 3787–3792.
Boucher MJ, Morisset J, Vachon PH, Reed JC, Lainé J, Rivard N . MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. J Cell Biochem 2000; 79: 355–369.
Warr MR, Shore GC . Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med 2008; 8: 138–147.
Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW . MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 2004; 23: 5301–5315.
Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR . Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell 2006; 21: 749–760.
Zhong Q, Gao W, Du F, Wang X . Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005; 121: 1085–1095.
Sutherland C, Leighton IA, Cohen P . Inactivation of glycogen synthase kinase-3 beta by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem J 1993; 296: 15–19.
Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ . Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology 2005; 128: 2054–2065.
Haber DA, Settleman J . Cancer: drivers and passengers. Nature 2007; 446: 145–146.
Weinstein IB, Joe A . Oncogene addiction. Cancer Res 2008; 68: 3077–3080.
Shirasawa S, Furuse M, Yokoyama N, Sasazuki T . Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 1993; 260: 85–88.
Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F et al. B-RAF is a therapeutic target in melanoma. Oncogene 2004; 23: 6292–6298.
Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ et al. Oncogenic BRAFV600E inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res 2008 (in press).
Wickenden JA, Jin H, Johnson M, Gillings AS, Newson C, Austin M et al. Colorectal cancer cells with the BRAFV600E mutation are addicted to the ERK1/2 pathway for growth factor-independent survival and repression of BIM. Oncogene 2008 (in press).
Boisvert-Adamo K, Aplin AE . BRAF and PI3 kinase signaling protect melanoma cells from anoikis. Oncogene 2006; 25: 4848–4856.
Liu D, Liu Z, Jiang D, Dackiw AP, Xing M . Inhibitory effects of the mitogen-activated protein kinase kinase inhibitor CI-1040 on the proliferation and tumor growth of thyroid cancer cells with BRAF or RAS mutations. J Clin Endocrinol Metab 2007; 92: 4686–4695.
Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C . BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Mol Cancer Res 2008; 6: 751–759.
Eisenmann KM, VanBrocklin MW, Staffend NA, Kitchen SM, Koo HM . Mitogen-activated protein kinase pathway-dependent tumor-specific survival signaling in melanoma cells through inactivation of the proapoptotic protein bad. Cancer Res 2003; 63: 8330–8337.
Boisvert-Adamo K, Aplin AE . Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene 2008; 27: 3301–3312.
Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P . Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin Cancer Res 2007; 13: 4934–4942.
Sheridan C, Brumatti G, Martin SJ . Oncogenic B-RAFV600E inhibits apoptosis and promotes ERK-dependent inactivation of Bad and Bim. J Biol Chem 2008; 283: 22128–22135.
Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell 2005; 7: 227–238.
Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 2007; 129: 957–968.
Tetsu O, McCormick F . Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 2003; 3: 233–245.
Gysin S, Lee SH, Dean NM, McMahon M . Pharmacologic inhibition of RAF → MEK → ERK signaling elicits pancreatic cancer cell cycle arrest through induced expression of p27Kip1. Cancer Res 2005; 65: 4870–4880.
Plattner R, Gupta S, Khosravi-Far R, Sato KY, Perucho M, Der CJ et al. Differential contribution of the ERK and JNK mitogen-activated protein kinase cascades to Ras transformation of HT1080 fibrosarcoma and DLD-1 colon carcinoma cells. Oncogene 1999; 18: 1807–1817.
Keller JW, Franklin JL, Graves-Deal R, Friedman DB, Whitwell CW, Coffey RJ . Oncogenic KRAS provides a uniquely powerful and variable oncogenic contribution among RAS family members in the colonic epithelium. J Cell Physiol 2007; 210: 740–749.
Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ . BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007; 4: 1669–1679.
Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A . Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med 2007; 4: 1681–1689.
Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med 2007; 4: 1655–1667.
Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res 2007; 67: 11867–11875.
Janmaat ML, Kruyt FA, Rodriguez JA, Giaccone G . Response to epidermal growth factor receptor inhibitors in non-small cell lung cancer cells: limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt kinase pathways. Clin Cancer Res 2003; 9: 2316–2326.
Deininger MW, Goldman JM, Melo JV . The molecular biology of chronic myeloid leukaemia. Blood 2000; 96: 3343–3356.
Amarante-Mendes GP, McGahon AJ, Nishioka WK, Afar DE, Witte ON, Green DR . Bcl-2-independent Bcr/Abl-mediated resistance to apoptosis: protection in correlated with upregulation of Bcl-xL. Oncogene 1998; 16: 1383–1390.
Ravandi F, Kantajian HM, Talpaz M, O'Brien S, Faderl S, Giles FJ et al. Expression of apoptosis proteins in chronic myelogenous leukaemia: associations and significance. Cancer 2001; 91: 1964–1972.
Aichberger KJ, Mayerhofer M, Krauth MT, Skvara H, Florian S, Sonneck K et al. Identification of mcl-1 as a Bcr/Abl-dependent target in chronic myeloid leukaemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonuceotides. Blood 2005; 105: 3303–3311.
Kuribara R, Honda H, Matsui H, Shinjyo T, Inukai T, Sugita K et al. Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors. Mol Cell Biol 2004; 24: 6172–6183.
Aichberger KJ, Mayerhofer M, Krauth MT, Vales A, Kondo R, Derdak S et al. Low level expression of proapoptotic Bcl-2-interacting mediator in leukemic cells in patients with chronic myeloid leukaemia: role of Bcr/Abl, characterization of underlying signalling pathways, and re-expression by novel pharmacologic compounds. Cancer Res 2005; 65: 9436–9444.
Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DCS et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukaemia cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci USA 2006; 103: 14907–14912.
Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A et al. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol 2001; 21: 854–864.
Nguyen TK, Rahamani M, Harada H, Dent P, Grant S . Mek1/2 inhibitors sensitize Bcr/Abl+ human leukaemia cells to the dual Abl/Src inhibitor BMS-345/825. Blood 2007; 109: 4006–4015.
Hüser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, Giblett S et al. MEK kinase activity is not necessary for Raf-1 function. EMBO J 2001; 20: 1940–1951.
Garnett MJ, Rana S, Paterson H, Barford D, Marais R . Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell 2005; 20: 963–969.
Tran SE, Holmstrom TH, Ahonen M, Kahari VM, Eriksson JE . MAPK/ERK overrides the apoptotic signaling from Fas, TNF, and TRAIL receptors. J Biol Chem 2001; 276: 16484–16490.
Acknowledgements
We apologise to colleagues whose work we have had to omit because of space constraints. We thank members of the Cook group past and present for advice over the years. We are also grateful to Paul Smith for many thoughtful discussions. Work in the author's laboratory is funded by the Association for International Cancer Research, AstraZeneca, the Biotechnology and Biological Sciences Research Council and The Babraham Institute.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by A Verkhratsky
Rights and permissions
This article is cited by
-
Protein interaction networks characterizing the A549 cells Klotho transfected are associated with activated pro-apoptotic Bim and suppressed Wnt/β-catenin signaling pathway
Scientific Reports (2024)
-
ROS production by mitochondria: function or dysfunction?
Oncogene (2024)
-
Proteomic analysis reveals the molecular mechanism of Astragaloside in the treatment of non-small cell lung cancer by inducing apoptosis
BMC Complementary Medicine and Therapies (2023)
-
MAPK-negative feedback regulation confers dependence to JAK2V617F signaling
Leukemia (2023)
-
A biscarbene gold(I)-NHC-complex overcomes cisplatin-resistance in A2780 and W1 ovarian cancer cells highlighting pERK as regulator of apoptosis
Cancer Chemotherapy and Pharmacology (2023)