Abstract
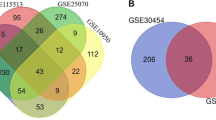
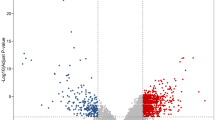
Colorectal cancer (CRC) is one of the most common malignant tumor and prevalent cause of cancer-related death worldwide. In this study, we analyzed the gene expression profiles of patients with CRC with the aim of better understanding the molecular mechanism and key genes in CRC. Four gene expression profiles including, GSE9348, GSE41328, GSE41657, and GSE113513 were downloaded from GEO database. The data were processed using R programming language, in which 319 common differentially expressed genes including 94 up-regulated and 225 down-regulated were identified. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analyses were conducted to find the most significant enriched pathways in CRC. Based on the GO and KEGG pathway analysis, the most important dysregulated pathways were regulation of cell proliferation, biocarbonate transport, Wnt, and IL-17 signaling pathways, and nitrogen metabolism. The protein–protein interaction (PPI) network of the DEGs was constructed using Cytoscape software and hub genes including MYC, CXCL1, CD44, MMP1, and CXCL12 were identified as the most critical hub genes. The present study enhances our understanding of the molecular mechanisms of the CRC, which might potentially be applied in the treatment strategies of CRC as molecular targets and diagnostic biomarkers.





Similar content being viewed by others
Data availability
All relevant data are within the paper.
References
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64.
Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017;18(4):722.
Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33(10):111.
Lv J, Li L. Hub genes and key pathway identification in colorectal cancer based on bioinformatic analysis. BioMed Res Int. 2019;2019:1545680.
Mohapatra SK, Krishnan A. Microarray data analysis. Plant reverse genetics. Totowa: Humana Press; 2011. p. 27–43.
Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15.
Smyth GK. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. p. 397–420.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–52.
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4(1):2.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–58.
Sakai E, Fukuyo M, Ohata K, Matsusaka K, Doi N, Mano Y, et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int J Cancer. 2016;138(7):1634–44.
Lu W, Li N, Liao F. Identification of key genes and pathways in pancreatic cancer gene expression profile by integrative analysis. Genes. 2019;10(8):612.
Huang Q, Wu L-Y, Wang Y, Zhang X-S. GOMA: functional enrichment analysis tool based on GO modules. Chin J Cancer. 2013;32(4):195.
Perez R, Wu N, Klipfel AA, Beart RW. A better cell cycle target for gene therapy of colorectal cancer: cyclin G. J Gastrointest Surg. 2003;7(7):884–9.
Tominaga O, Nita ME, Nagawa H, Fujii S, Tsuruo T, Muto T. Expressions of cell cycle regulators in human colorectal cancer cell lines. Jpn J Cancer Res. 1997;88(9):855–60.
Djamgoz MB, Coombes RC, Schwab A. Ion transport and cancer: from initiation to metastasis. London: The Royal Society; 2014.
Han B, Feng D, Yu X, Zhang Y, Liu Y, Zhou L. Identification and interaction analysis of molecular markers in colorectal cancer by integrated bioinformatics analysis. Med Sci Monit. 2018;24:6067.
Chen S, Wang Y, Zhang L, Su Y, Zhang M, Wang J, et al. Exploration of the mechanism of colorectal cancer metastasis using microarray analysis. Oncol Lett. 2017;14(6):6671–7.
Al-Rawi M, Rmali K, Watkins G, Mansel R, Jiang W. Aberrant expression of interleukin-7 (IL-7) and its signalling complex in human breast cancer. Eur J Cancer. 2004;40(4):494–502.
Wu D, Wu P, Huang Q, Liu Y, Ye J, Huang J. Interleukin-17: a promoter in colorectal cancer progression. Clin Dev Immunol. 2013;2013:436307.
Numasaki M, Watanabe M, Suzuki T, Takahashi H, Nakamura A, McAllister F, et al. IL-17 enhances the net angiogenic activity and in vivo growth of human non-small cell lung cancer in SCID mice through promoting CXCR-2-dependent angiogenesis. J Immunol. 2005;175(9):6177–89.
Benevides L, Cardoso CR, Tiezzi DG, Marana HR, Andrade JM, Silva JS. Enrichment of regulatory T cells in invasive breast tumor correlates with the upregulation of IL-17A expression and invasiveness of the tumor. Eur J Immunol. 2013;43(6):1518–28.
Li C, Zeng X, Yu H, Gu Y, Zhang W. Identification of hub genes with diagnostic values in pancreatic cancer by bioinformatics analyses and supervised learning methods. World J Surg Oncol. 2018;16(1):223.
Zhao ZW, Fan XX, Yang LL, Song JJ, Fang SJ, Tu JF, et al. The identification of a common different gene expression signature in patients with colorectal cancer. Math Biosci Eng. 2019;16(4):2942–58.
Liu J, Li H, Sun L, Shen S, Zhou Q, Yuan Y, et al. Epigenetic alternations of MicroRNAs and DNA methylation contribute to liver metastasis of colorectal cancer. Dig Dis Sci. 2019;64(6):1523–34.
Carter AB, Misyak SA, Hontecillas R, Bassaganya-Riera J. Dietary modulation of inflammation-induced colorectal cancer through PPAR. PPAR Res. 2009;2009:498352.
Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39(6):527–79.
Wang Z, Tang Y, Xie L, Huang A, Xue C, Gu Z, et al. The prognostic and clinical value of CD44 in colorectal cancer: a meta-analysis. Front Oncol. 2019;9:309.
Du L, Wang H, He L, Zhang J, Ni B, Wang X, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14(21):6751–60.
Bourguignon LY, Singleton PA, Diedrich F, Stern R, Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J Biol Chem. 2004;279(26):26991–7007.
Harada N, Mizoi T, Kinouchi M, Hoshi K, Ishii S, Shiiba K, et al. Introduction of antisense CD44s cDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91(1):67–75.
Murray GI, Duncan ME, O’Neil P, Melvin WT, Fothergill JE. Matrix metalloproteinase–1 is associated with poor prognosis in colorectal cancer. Nat Med. 1996;2(4):461–2.
Wang Q, Chen S, Wu J, Liu D, Jiang N, Wang B, et al. Identification of potential hub genes and signal pathways promoting the distinct biological features of cord blood-derived endothelial progenitor cells via bioinformatics. Genet Test Mol Biomark. 2020. https://doi.org/10.1089/gtmb.2019.0272.
Melhem MF, Meisler AI, Finley GG, Bryce WH, Jones MO, Tribby II, et al. Distribution of cells expressing myc proteins in human colorectal epithelium, polyps, and malignant tumors. Cancer Res. 1992;52(21):5853–64.
Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci. 2017;114(37):E7697–706.
Gu L, Chu P, Lingeman R, McDaniel H, Kechichian S, Hickey RJ, et al. The mechanism by which MYCN amplification confers an enhanced sensitivity to a PCNA-derived cell permeable peptide in neuroblastoma cells. EBioMedicine. 2015;2(12):1923–31.
Yuan M, Zhu H, Xu J, Zheng Y, Cao X, Liu Q. Tumor-derived CXCL1 promotes lung cancer growth via recruitment of tumor-associated neutrophils. J Immunol Res. 2016. https://doi.org/10.1155/2016/6530410.
Kong D-J, Qin Y-F, Li G-M, Zhao Y-M, Hao J-P, Wang H. Identification of hub gene TIMP1 and relative ceRNAs regulatory network in colorectal cancer. 2020.
Hsu Y-L, Chen Y-J, Chang W-A, Jian S-F, Fan H-L, Wang J-Y, et al. Interaction between tumor-associated dendritic cells and colon cancer cells contributes to tumor progression via CXCL1. Int J Mol Sci. 2018;19(8):2427.
le Rolle A-F, Chiu TK, Fara M, Shia J, Zeng Z, Weiser MR, et al. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. J Trans Med. 2015;13(1):1–12.
Nagasawa T. CXC chemokine ligand 12 (CXCL12) and its receptor CXCR4. J Mol Med. 2014;92(5):433–9.
Wendt MK, Johanesen PA, Kang-Decker N, Binion DG, Shah V, Dwinell MB. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene. 2006;25(36):4986–97.
Chen L, Pan X, Hu X, Zhang YH, Wang S, Huang T, et al. G ene expression differences among different MSI statuses in colorectal cancer. Int J Cancer. 2018;143(7):1731–40.
Ma Y-S, Huang T, Zhong X-M, Zhang H-W, Cong X-L, Xu H, et al. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol Cancer. 2018;17(1):139.
Zhang G-L, Pan L-L, Huang T, Wang J-H. The transcriptome difference between colorectal tumor and normal tissues revealed by single-cell sequencing. J Cancer. 2019;10(23):5883.
Zhang TM, Huang T, Wang RF. Cross talk of chromosome instability, CpG island methylator phenotype and mismatch repair in colorectal cancer. Oncol Lett. 2018;16(2):1736–46.
Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330.
Li B-Q, Huang T, Liu L, Cai Y-D, Chou K-C. Identification of colorectal cancer related genes with mRMR and shortest path in protein-protein interaction network. PLoS ONE. 2012;7(4):e33393.
Jiang Y, Huang T, Chen L, Gao Y-F, Cai Y, Chou K-C. Signal propagation in protein interaction network during colorectal cancer progression. BioMed Res Int. 2013;2013:287019.
Li B-Q, Huang T, Zhang J, Zhang N, Huang G-H, Liu L, et al. An ensemble prognostic model for colorectal cancer. PLoS ONE. 2013;8(5):e63494.
Zheng Z, Xie J, Xiong L, Gao M, Qin L, Dai C, et al. Identification of candidate biomarkers and therapeutic drugs of colorectal cancer by integrated bioinformatics analysis. Med Oncol. 2020;37(11):104. https://doi.org/10.1007/s12032-020-01425-2.
Author information
Authors and Affiliations
Contributions
HH designed the study. HH and S-MR implemented the methods and analyzed the data. HH, AL contributed to the interpretation of the results. HH, S-MR, AL drafted the manuscript and prepared all figures and tables. AM outlined the methods and edited the manuscript and figures. All authors participated in improving the writing of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hozhabri, H., Lashkari, A., Razavi, SM. et al. Integration of gene expression data identifies key genes and pathways in colorectal cancer. Med Oncol 38, 7 (2021). https://doi.org/10.1007/s12032-020-01448-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-020-01448-9